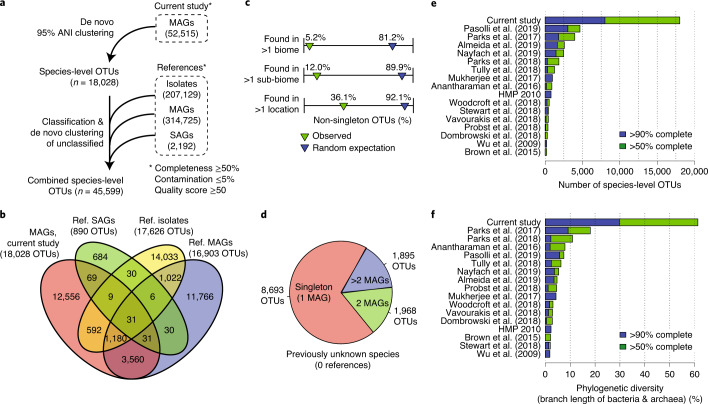
Fig. 2. Species-level clustering of the GEM catalog with >500,000 reference genomes.
a, MAGs from the current study were compared to 524,046 publicly available reference genomes found in IMG/M and NCBI. All reference genomes met the same minimum quality standards as applied to the GEM catalog. All MAGs and reference genomes were clustered into 45,599 species-level OTUs on the basis of 95% ANI and 30% AF. b, Overlap of OTUs between genome sets. MAGs from the current study revealed genomes for 12,556 species for the first time. c, The vast majority of OTUs with >1 genome from the GEM catalog were restricted to individual biomes and sub-biomes, although over a third were found in multiple geographic locations. d, A large proportion of the 12,556 newly identified species were represented by only a single genome. e,f, Comparison of the current dataset with the 16 largest previously published genome studies, selected on the basis of species-level diversity. Study identifiers were derived from either NCBI BioProject or GOLD. Studies by Wu et al. 35, HMP (2010)36 and Mukherjee at al. 34 contain additional genomes generated after publication. All MAGs from other studies were filtered using the same quality criteria as the GEM dataset (Fig. 1a and Methods). Genomes from the current study represent over three times more diversity compared to any previously published study.