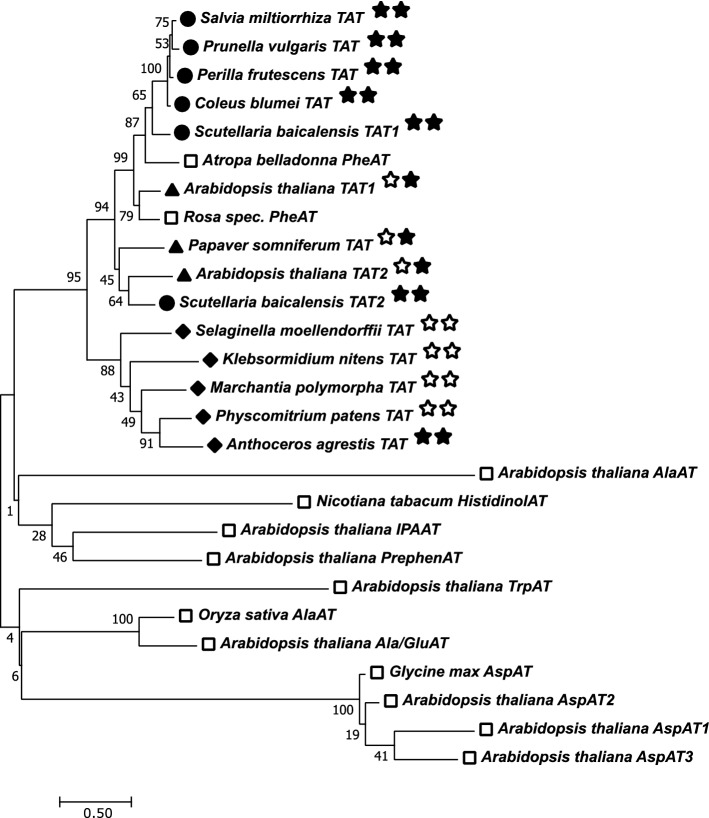
Fig. 7.
Phylogenetic tree (maximum likelihood analysis) of TAT amino acid sequences of several members of the Lamiaceae (circles) and representatives of other higher plant families (triangles) and lower plants (diamonds). Analyses were conducted in Mega 7 (Kumar et al. 2016). The tree with the highest log likelihood (− 13,478.93) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (bootstrap replicates 1000). Underlying TAT amino acid sequences: Salvia miltiorrhiza (ABC60050), Prunella vulgaris (AJW87632), Perilla frutescens (ADO17550), Coleus blumei (CAD30341), Scutellaria baicalensis (AIV98132 for SbTAT1 and AIV98133 for SbTAT2), Arabidopsis thaliana (NP_200208 for AtTAT1 and NP_198465 for AtTAT2), Papaver somniferum (ADC33123), Selaginella moellendorffii (XP_024536669), Klebsormidium nitens (GAQ85880), Marchantia polymorpha (OAE18929), Physcomitrium patens (XP_024361307), Anthoceros agrestis (MN922307, this paper). The two stars following the name indicate the presence of RA in the plant (first star) and an experimentally proven TAT activity of the underlying protein (second star), if filled in. Underlying amino acid sequences of distantly related aminotransferases (open squares): Arabidopsis thaliana TrpAT (AAO63403), A. thaliana IPAAT (AAP68293), A. thaliana PrephenAT (Q9SIE1), Oryza sativa AlaAT (BAA77261), A. thaliana AlaAT (Q9SR86), A. thaliana Ala/GluAT (AAN62333), Glycine max AspAT (AAC50015), A. thaliana (mitochondrial) AspAT1 (P46643), A. thaliana (cytoplasmic) AspAT2 (P46645), A. thaliana (plastidic) AspAT5 (P46248), Nicotiana tabacum HistidinolAT (CAA70403), Atropa belladonna PheAT (AHN10104), Rosa ‘Yves Piaget’ PheAT (Hirata et al. 2012)