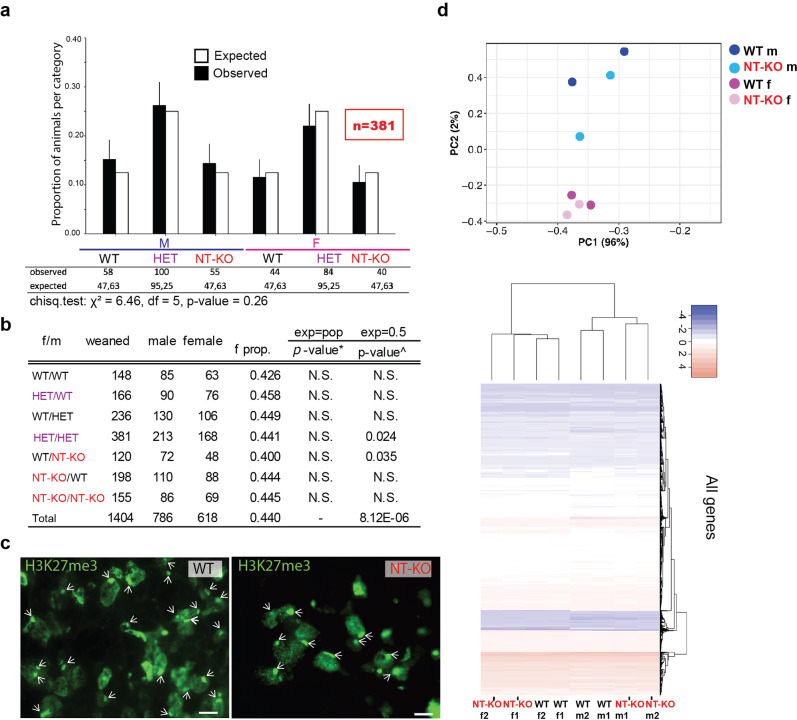
Fig. 2. A N-term LBR deletion, does not affect XCI in vivo.
a 34 Lbr heterozygous (Lbr+/Lbr236, HET) mice pairs were crossed and the offspring have been genotyped by PCR analysis (see schematic in Supplementary Fig. 1a). Observed offspring genotypes indicated in black with error bars indicating 95% confidence interval (CI). A significant skewing against females is predicted in the analyzed offspring, in case Lbr plays a major role in XCI in vivo (M: male; F: female). Chi-square test analysis reveals no significant differences between expected and observed classes of male and female offspring (X2 = 6.46, df = 5, p-value = 0.26). The number of animals used is shown in red (n = 381). b The table shows different classes of crosses and indicates the parental origin and type of the mutation. Sex ratio and binomial test analysis are also shown. Statistical tests are run vs the observed total population ratio of males/females (m/f) and vs the expected, theoretical 50% ratio. *Two-tailed binomial test against population female proportion (0.440). ^Two-tailed binomial test against expected female proportion (0.50). c Xi-lamina association reveals no differences between WT and LBR NT-KO animals. H3K27me3 staining (Xi surrogate) is shown. Cross-sections from thymus tissue are shown. Arrows indicate the inactive X chromosome. Scale bar indicates 10 µm. d Top, RNA-Seq principal component analysis and hierarchical clustering of the LBR NT-KO vs WT are shown (number of genes: 12,791). The heatmap depicts scaled expression for better visualization. Categories are shown in the legends.