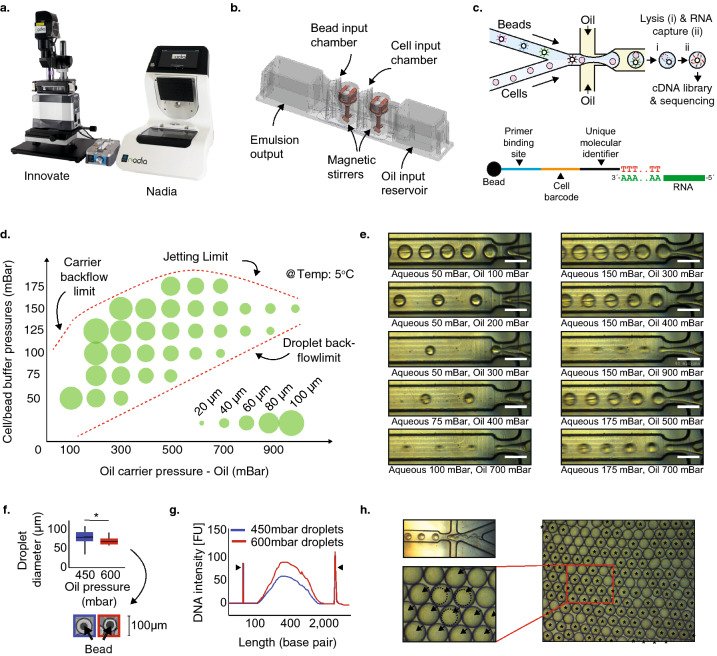
Figure 1.
An open platform for single cell transcriptome profiling. (a) The Nadia Instrument (right) and Nadia Innovate (left) benchtop platform for single-cell transcriptomics. (b) Design of the disposable microfluidics cartridge used in the Nadia. (c) Schematic of the droplet sequencing workflow used in the Nadia platform. In brief, single cells or nuclei are encapsulated in aqueous droplets in oil together with barcoded beads. Following lysis within droplets the released mRNA is captured upon the bead and provided both a cell barcode and a unique molecular identifier. Beads are subsequently pooled prior to reverse-transcription and generation of cDNA libraries called “single-cell transcriptomes attached to microparticles” (STAMPs). The barcoded STAMPs are then amplified in pools for high-throughput RNA-seq. (d) Theoretical variation of droplet size by changing oil and liquid stream pressures. (e) Experimental variation of droplet size by changing oil and liquid stream pressures. White scale bars represent 100 μm. (f) Stable droplet diameters at different oil pressures. Inset shows example droplets containing non-deformable beads. (g) Bioanalyser traces of full-length transcript PCRs amplified from identical bead numbers but different droplet dimensions. (h) Example image of deformable beads captured with the Nadia system. Upper left panel shows crowding of deformable beads behind microfluidics junction, lower left panel shows droplet occupancy following sychronised deformable bead loading. For reader guidance, outlines of three deformable beads are indicated with dashed lines, and droplets containing beads are marked by black arrowheads. Right panel shows zoomed out image revealing > 70% droplet occupancy of deformable beads. For reader guidance, all droplets containing a deformable bead are marked by a black asterik.