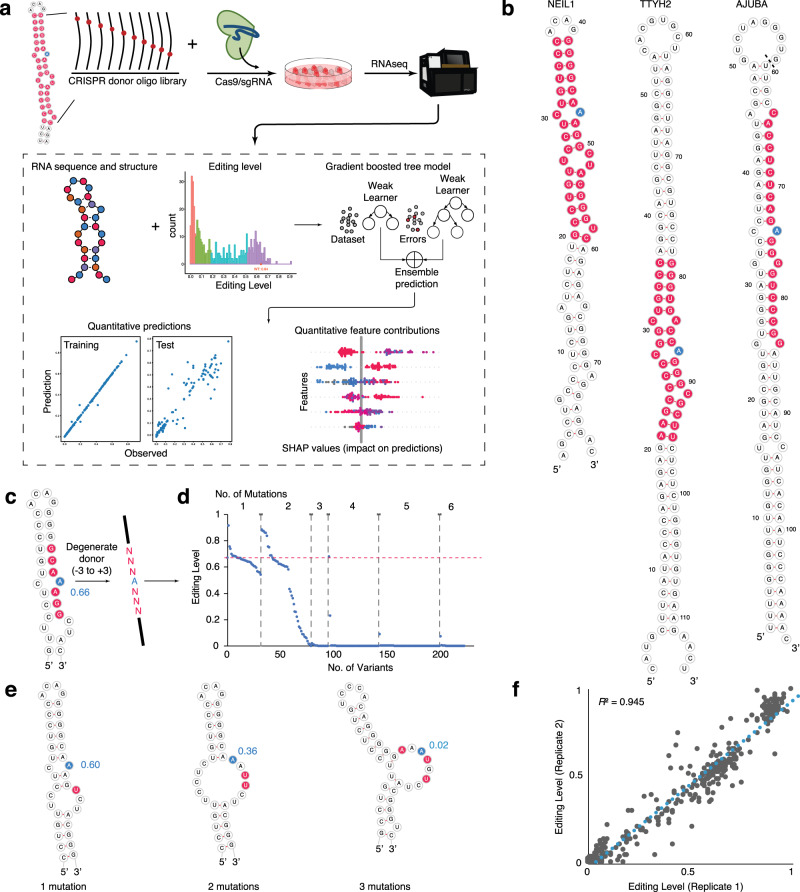
Fig. 1. CRISPR/Cas9-mediated mutagenesis in endogenous RNA to dissect RNA editing by ADAR1 in cells.
a Overview of the experimental methods and computational pipeline. CRISPR/Cas9-mediated homology-directed repair is applied to mutagenesis of endogenous RNA in HEK293T cells. A supervised machine learning method (a gradient boosted tree, XGBoost) was applied to develop quantitative models that predict how cis-elements, such as RNA sequence and secondary structure determine RNA editing level. b Sequence and secondary structure of the three RNAs, NEIL1, TTYH2, and AJUBA, for targeted mutagenesis. The residues subjected to mutations are highlighted in red and the specific editing site is in blue. For AJUBA, partial sequences from the genomic sequences are taken to focus on the region of interest. Therefore, the G59 and U60 shown in b is 524 nt apart in the genomic region. c Degenerate donor oligos are designed for the −3 to +3 nt region around the specific editing site in the NEIL1 substrate. The mutagenized region is highlighted in red and the editing site in blue. The value of editing level is shown in blue. d The distribution of editing level by the number of mutations from the results of the degenerate NEIL1 library from c. e Examples of how the number of mutations affect the RNA secondary structure of NEIL1. The mutagenized nucleotied is highlighted in red and the editing site in blue. The value of editing level is shown in blue. f Reproducible editing measurement of the two replicates of the targeted mutagenesis library of NEIL1 shown by pairwise comparison with Spearman R2 labeled.