Figure 4.
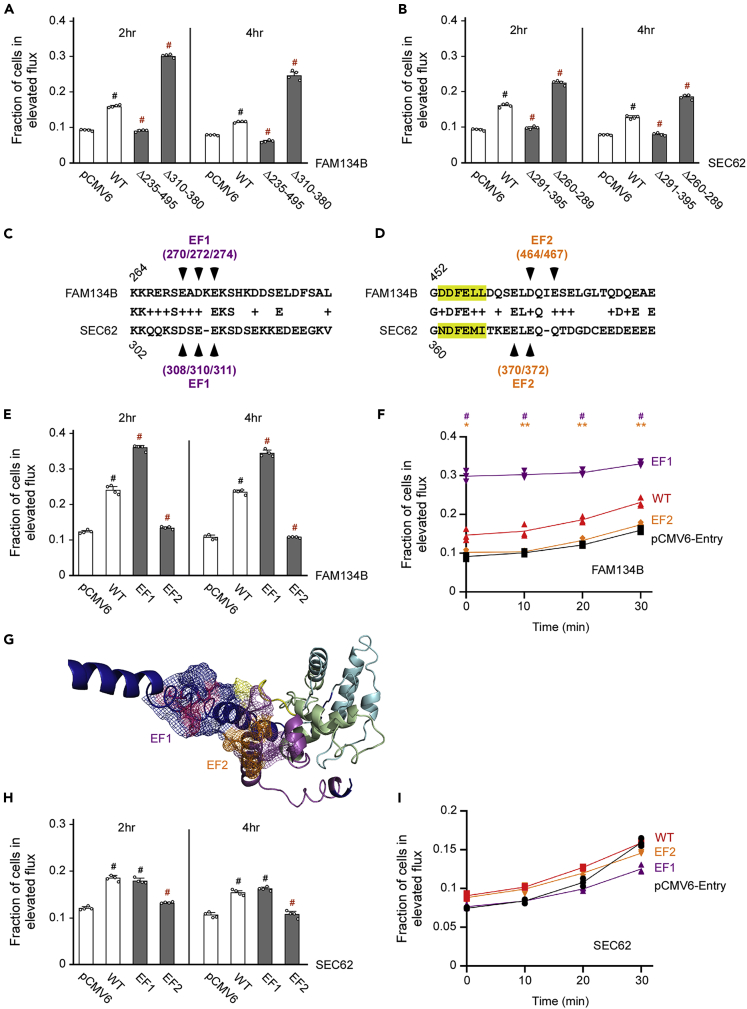
FAM134B and SEC62 mutants that engage ATG9A can induce reticulophagy
(A and B) Reticulophagy flux induced by different FAM134B (A) or SEC62 (B) deletion mutants. Data are represented as mean ± SD. #p < 0.0005.
(C and D) Sequence alignment of presumed EF hands in FAM134B and SEC62. Both EF1 (C) and EF2 (D) are shown. The Ca++-stabilizing amino acids that were mutated in the following experiments are indicated by arrowheads. The LIR domain is highlighted.
(E and F) EF1 and EF2 of FAM134B differentially affect reticulophagy flux. (E) Flux at 2hr and 4hr. (F) Time-course. Data are represented as mean ± SD. ∗p < 0.05; ∗∗p < 0.005; #p < 0.0005.
(G) I-TASSER model of the opposing EF1 and EF2 of FAM134B showing predicted interaction.
(H and I) EF1 and EF2 of SEC62 have a modest effect on the induction of reticulophagy. (H) Flux at 2hr and 4hr. (I) Time-course. Data are represented as mean ± SD. #p < 0.0005.