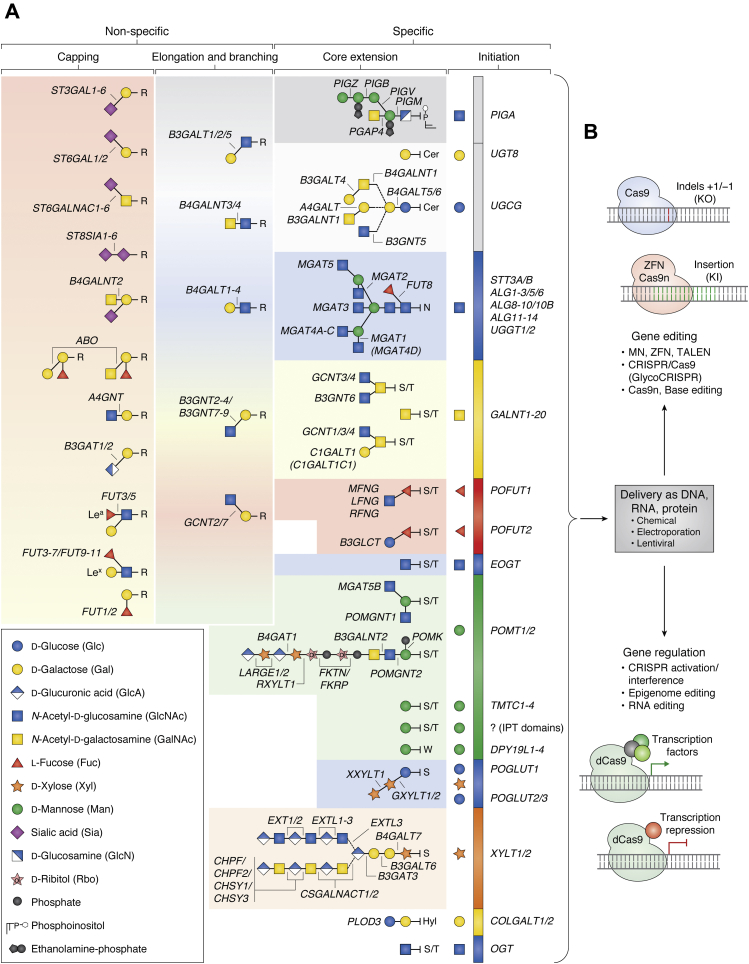
Figure 2.
Overview of principles and strategies for stable genetic engineering of cellular glycosylation capacities.A, overview of the 16 human glycosylation pathways with predicted assignments of 173 glycosyltransferase genes to the major biosynthetic steps using the rainbow display organization (5, 199). The rainbow depiction of glycosylation pathways illustrates the major biosynthetic steps organized into pathway-specific steps (right part with even colored according to the initial monosaccharide except for glycolipids) and pathway nonspecific steps (left part with toned colors) with predicted GT genes assigned. Note that this is a simplified scheme of pathways and GT genes are assigned only to the primary predicted functions. Genetic engineering of glycosylation requires considering the properties of individual enzymes and their potential effects on the cellular glycosylation pathways. Loss or gain of a GT may have highly specific effects or wider effects on multiple glycosylation pathways. Glycosylation steps covered exclusively by one unique enzyme (nonredundant steps), e.g., core α6-fucosylation of N-glycans by FUT8, yield highly specific and predictable outcomes with loss/gain engineering. Steps covered by multiple isoenzymes with overlapping functions (redundant steps) may or may not yield easily predictable outcomes, and the outcome may vary in cells dependent on the expression of such isoenzymes. Most steps in elongation and branching and capping are covered by partial redundancies by multiple isoenzymes. For example, sialylation by any of the four sialyltransferase subfamilies is covered by partial redundancies, and, e.g., combinatorial KO of three genes is required to selectively eliminate α3-sialylation on N-glycans (KO of ST3GAL3/4/6), while KO of two genes is required to selectively eliminate α3-sialylation of core1 O-glycans (KO ST3GAL1/2). Glycan symbols are displayed in the Symbol Nomenclature for Glycans (SNFG) format (340). B, graphic depiction of current nuclease-based gene-editing tools for knockout (KO) and knock-in (KI) of genes, and emerging CRISPR-based technologies for regulating and activating gene expression.