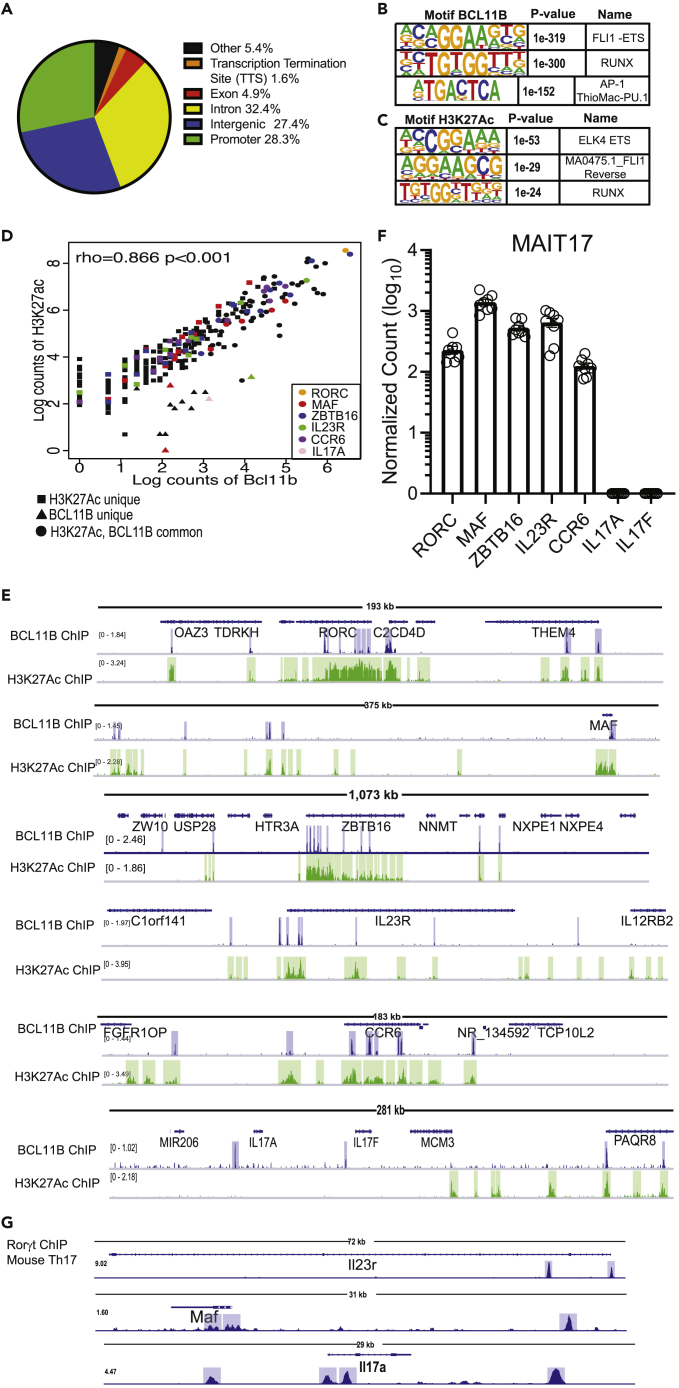
Figure 6.
BCL11B binds at H3K27Ac-rich regions of key genes of MAIT17 program in human MAIT cells from healthy donors
(A) Peak distribution of BCL11B binding sites genome-wide in human MAIT cells sorted as CD3+CD161+TCRVα7.2+ from PBMCs of healthy donors. (B) Top 3 BCL11B binding motifs in human MAIT cells.
(C) Top 3 BCL11B motifs overlapping with H3K27Ac peaks. CD3+CD161+TCRVα7.2+ were sorted from PBMCs derived from nine donors (BCL11B ChIP-seq) and two donors (H3K27Ac ChIP-seq).
(D) Spearman's Correlation plot of BCL11B binding intensity with H3K27Ac peaks at MAIT17 signature genes. Several genes of the MAIT17 program are listed.
(E) Integrative Genomics Viewer (IGV) visualization at RORC, MAF, ZBTB16, IL23R, CCR6 and IL17A and IL17F loci of BCL11B ChIP-seq (track 1) and H3K27Ac ChIP-seq (track 2) in human MAIT cells sorted from PBMCs of healthy donors.
(F) Normalized counts in log10 format for RORC, MAF, ZBTB16, IL17A, IL17F, IL23R, CCR6 in blood MAIT cells of healthy donors. Data analyzed from (Dias et al., 2018). n = 8. Data are represented as mean ± SEM.
(G) IGV visualizations of Rorγt ChIP-seq in murine Th17 cells at IL23r, Maf, and IL17a loci. Data analyzed from (Ciofani et al., 2012). Blue and green highlights indicate significant ChIP-seq peaks. ChIP-seq scale bar normalized to sequences per million reads.