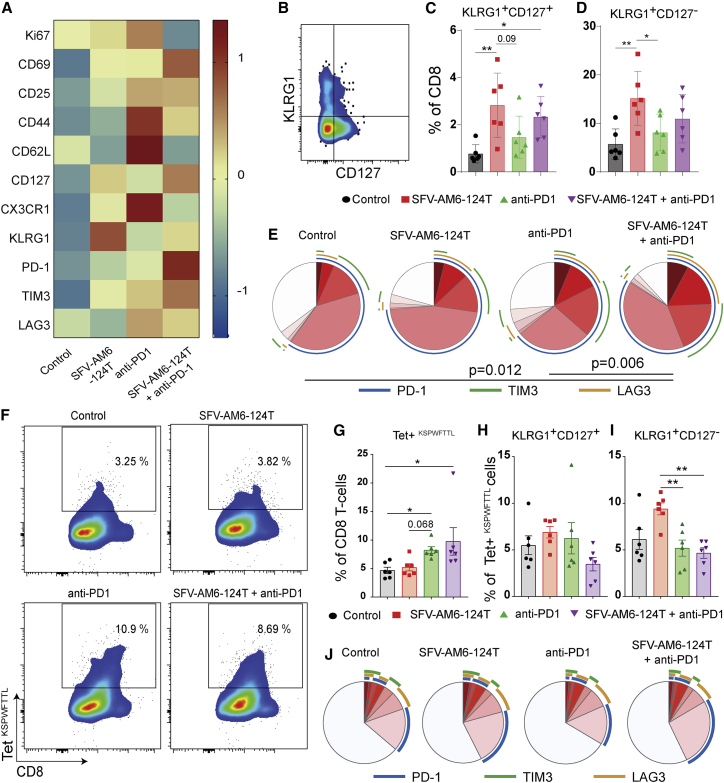
Figure 5.
SFV-AM6-124T promotes a distinct CD8+ T cell phenotype in GL261 gliomas
(A) MFI heatmap of Ki67, CD69, CD25, CD44, CD62L, CD127, CX3CR1, KLRG1, PD1, TIM3, and LAG3 expression in CD8+ T cells from control and treated tumors. (B) Flow cytometry gating strategy for quantification of KLRG1 and CD127-expressing CD8+ T cells. (C and D) Quantification of KLRG1+/CD127− (C) and KLRG1+/CD127+ (D) cells. (E) Quantification of PD-1, TIM3, and LAG3 co-expression on CD8 T cells with SPICE software. Colors of the pie arcs depict the expression of individual inhibitory receptors, while the pie depicts the average proportion of co-expressed inhibitory receptors. (F) Bivariate plots showing staining for tetramer (Kb-restricted peptide amino acids 604–611 of p15E protein [KSPWFTTL]) on tumor-infiltrating CD8 T cells, and (G) the quantification of tumor-infiltrating tetramer+ CD8 T cells in each treatment group. (H and I) Quantification of KLRG1+/CD127− (H) and KLRG1+/CD127+ (I) tetramer+ CD8 T cell subpopulation. (J) Quantification of PD-1, TIM3, and LAG3 co-expression on tetramer+ CD8 T cells with SPICE software. Panels C, D, G, H and I are plotted as mean ± SD. Statistical analysis was done with one-way ANOVA with Tukey’s post hoc test (C, D, H and I) or Kruskal-Wallis with Dunn's post hoc test (G). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.