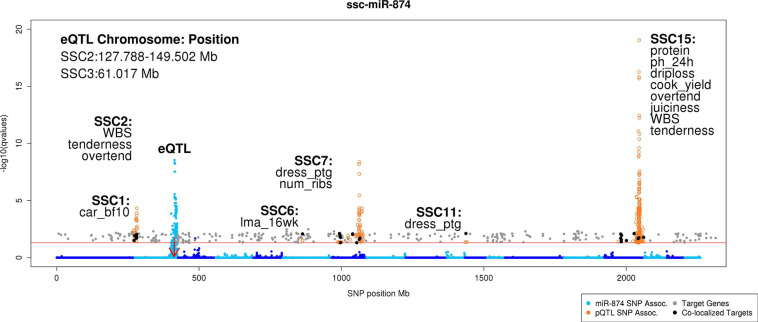
FIGURE 4.
Manhattan plot of genomic colocalization events of miR-874 target genes with pQTL. This Manhattan plot compiles the results of the miR-eQTL, previously identified mRNA eQTL (Velez-Irizarry et al., 2019), previously identified pQTL (Gualdrón Duarte et al., 2016; Casiró et al., 2017; Velez-Irizarry et al., 2019), miR-874 target prediction, and correlation analyses to identify potential novel mechanisms regulating growth, carcass composition, and meat quality traits in pigs. In total, 29 unique target genes colocalized with pQTL across six chromosomes. Nine target genes (PLEKHB2, WDR33, NIF3L1, IGFBP5, UGGT1, PRPF40A, METTL8, PTPRN, and KLHL30) overlap the pQTL peak on SSC15 associated with meat quality traits. The y-axis denotes the significance of the association [-log10(q value)], and the x-axis indicates the absolute SNP position (Mb). Chromosomes are differentiated by shades of blue. Blue points represent miRNA-SNP associations for miR-874. The red arrow indicates the genomic position of the miR-874 precursor transcript. Orange circles represent the genomic position of pQTL. Gray points represent the genomic position of significantly negatively correlated target genes for miR-874. Black points represent the genomic position of significantly negatively correlated target genes for miR-874 whose genomic position overlaps that of a pQTL peak. Text in the upper left-hand side of plot describes the genomic position of the miR-eQTL peaks (Mb). Text in close proximity to pQTL peaks colocalizing with target genes describe the chromosome on which the pQTL resides and which phenotypes the associated pQTL represents (car_bf10, carcass 10th rib backfat thickness; cook_yield, cook yield; dress_ptg, dressing percentage; driploss, driploss; juiciness, sensory panel juiciness; lma_16wk, loin muscle area at 16 weeks of age; num_ribs, number of ribs; overtend, sensory panel overall tenderness; ph_24h, pH measured at 24 h post-mortem; protein, protein percentage; tenderness, sensory panel tenderness; WBS, Warner Bratzler shear force).