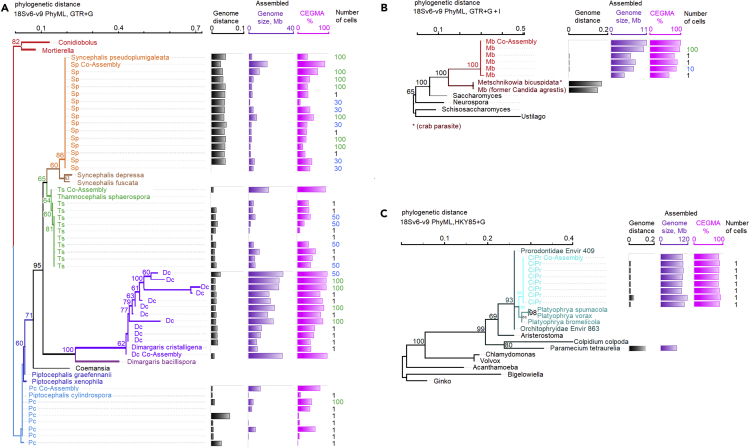
Figure 6.
Intraspecific single-cell genome variability and phylogenetic placement of single- and multi-cell genomes
Phylogenetic distance was estimated based on the 18S rDNA region v6 through v9 using PhyML package (Guindon et al., 2010). Genome distance was estimated using Genome-to-Genome Distance Calculator (GGDC), formula 2 (Meier-Kolthoff et al., 2013). See Table 3 for Genome-to-Genome Distance between genera.
(A) Zoopagomycota phylogenetic tree and GGDC, genome size, and completeness. Best nucleotide substitution model estimated HKY85, random starting tree, estimated best tree with bootstrap analysis, bootstrap shown values above 60%. Tree: Branches are shown as: Dc, Dimargaris cristalligena RSA 468; Ts, Thamnocephalis sphaerospora, Sp, Syncephalis pseudoplumigaleata; Pc, Piptocephalis cylindrospora RSA2659.
(B) Ascomycota single-cell phylogenetic tree and GGDC, genome size, and completeness. Best nucleotide substitution model estimated GTR+G+I, random starting tree, estimated best tree with bootstrap analysis, bootstrap shown values above 60%. Tree: Branches are shown as Mb, Metschnikowia bicuspidata in red with closest species in dark red.
(C) Compost ciliate single-cell phylogenetic tree and GGDC, genome size and completeness. Best nucleotide substitution model estimated HKY85+G, with bootstrap analysis, shown values above 60%. Branches for the single-cells are shown with: aqua, ciliate Protist (CiPr). Species with closest 18S are shown in dark teal. Non-Alveolata branches are shown in black.