Abstract
Plant growth promoting rhizobacteria (PGPR) can display several plant-beneficial properties, including support to plant nutrition, regulation of plant growth, and biocontrol of pests. Mechanisms behind these effects are directly related to the presence and expression of specific genes, and different PGPR strains can be differentiated by the presence of different genes. In this study we reported a comprehensive evaluation of a novel PGPR Klebsiella variicola UC4115 from the field to the lab, and from the lab to the plant. The isolate from tomato field was screened in-vitro for different activities related to plant nutrition and growth regulation as well as for antifungal traits. We performed a functional annotation of genes contributing to plant-beneficial functions previously tested in-vitro. Furthermore, the in-vitro characterization, the whole genome sequencing and annotation of K. variicola UC4115, were compared with the well-known PGPR Azospirillum brasilense strain Sp7. This novel comparative analysis revealed different accumulation of plant-beneficial functions contributing genes, and the presence of different genes that accomplished the same functions. Greenhouse assays on tomato seedlings from BBCH 11–12 to BBCH > 14 were performed under either organic or conventional management. In each of them, three PGPR inoculations (control, K. variicola UC4115, A. brasilense Sp7) were applied at either seed-, root-, and seed plus root level. Results confirmed the PGP potential of K. variicola UC4115; in particular, its high value potential as indole-3-acetic acid producer was observed in increasing of root length density and diameter class length parameters. While, in general, A. brasilense Sp7 had a greater effect on biomass, probably due to its high ability as nitrogen-fixing bacteria. For K. variicola UC4115, the most consistent data were noticed under organic management, with application at seed level. While, A. brasilense Sp7 showed the greatest performance under conventional management. Our data highlight the necessity to tailor the selected PGPR, with the mode of inoculation and the crop-soil combination.
Keywords: Klebsiella variicola, Azospirillum brasilense, PGPR, genome analyses, inoculation
Introduction
Plant growth promoting rhizobacteria (PGPR) are a heterogenous group of soil-dwelling bacteria able to efficiently colonize plants root system, enhancing plant nutrition, stress tolerance and health (Vacheron et al., 2013). PGPR can improve crops fitness either directly and indirectly. Direct processes include the promotion of alternative nutrient uptake pathway, the solubilization or fixation of chemical nutrients, and the production of various phytohormones; on the other hand indirect processes reduce or prevent the deleterious effects of one or more phytopathogenic organisms, through the synthesis of numerous antimicrobial compounds, and/or inducing systemic resistance (Oleńska et al., 2020). Since the recognition of their capabilities, several hundreds of candidate PGPR strains have been screened and evaluated in laboratory, greenhouse and field studies across the world (Martínez-Viveros et al., 2010). However, despite extensive literature on the mode of actions of PGPR, the implementation of this biotechnology in the agriculture industry, has been limited due to the variability information of the effectiveness of PGPR on plant growth, which could influence crop production (Vejan et al., 2016).
In recent years, the genome characterization of emblematic PGPR model strains have uncovered the molecular basis of some of their beneficial effects, leading the identification of genes that are involved in the PGPR-plant cooperation (Bruto et al., 2014; Liu et al., 2016; Shariati et al., 2017). According to these studies, many PGPR strains are multifunctional, i.e., they harbor more than one plant-beneficial properties, and this suggests the presence of conservative genes commonly distributed among different genera of microorganisms. However, a recent study based on ancestral character reconstruction, has proposed that PGPR-plant cooperation may have established separately in various taxa, yielding PGPR strains that use different gene assortments (Bruto et al., 2014). A holistic comparative discussion between in-vitro and in-vivo assay, and genomics could enhance the observation how these different assortments of plant-beneficial functions contributing genes (PBFC genes) affect the eukaryotic host.
A concrete observation about contribution of different multifunctional PGPR, can be performed using greenhouse horticultural crop, since different promotion capacity can be more easily determined under semi-controlled conditions (Ruzzi and Aroca, 2015). So far, organic production of high-value horticulture is one of the most important areas of PGPR application (Olowe et al., 2020). Tomato (Solanum lycopersicum L.) is one of the most demanding high-value horticultural crops worldwide, after potato (Singh et al., 2018), and is a good crop model study for PGPR effectiveness, because in the majority of cases, inoculation with PGPR always determined an increase in plant root length and plant productivity (Ruzzi and Aroca, 2015).
The genus Klebsiella, is one of the common genera detected in the rhizosphere system (Podile and Kishore, 2006), and the species Klebsiella variicola strains have been found to naturally colonize the rhizosphere of tomato plant (Guerrieri et al., 2020; Sunera et al., 2020). In particular, the plant growth-promoting (PGP) capacity of distinct strains of K. variicola were confirmed in glasshouse experiments as well as in field trials, both in terms of yield increase and product quality improvement (Kim et al., 2017; Yang and Yang, 2020). Plants seem to be the natural niches for this species, and its dominant feature is the capability to fix N2 (Chen et al., 2016). Actually, Klebsiella sp. are among the major free-living nitrogen fixing bacteria, together with Azospirillum brasilense, Pantoea agglomerans, Burkholderia sp., Serratia sp. (Bhattacharjee et al., 2008). Moreover, different PGP traits such as phosphorus solubilization, indole-3-acetic acid (IAA) production, and biocontrol activity were identified in distinct strains of K. variicola (Liu et al., 2014; Sekhar and Thomas, 2015). Despite the high PGP potential of this species, K. variicola is also known for its potential as opportunistic pathogens, with a number of strains of this species known to cause nosocomial respiratory and urinary tract infection, and pediatric outbreaks (Barrios-Camacho et al., 2019). A few K. variicola genomes analysis are currently reported, but the majority of investigations have focused on their pathogenic genes (Srinivasan and Rajamohan, 2020). By contrast, a shortage of studies is referred to the genes that contribute to the beneficial PGP activity of this species, except for nitrogen fixation.
Here, we used the high plant growth promoting potential of K. variicola strain UC4115 as a case study to present a comprehensive overview of an evaluating approach of PGPR: from the plant to the lab, through the phenotypic and genomic characterization of the isolate, and from the lab to the plant, through the direct observation of PGPR-plant beneficial cooperation. To obtain a better understanding of the effects of different PGPR on plant growth, the genomic and agronomical evaluation of K. variicola UC4115 was compared with that of Azospirillum brasilense Sp7, one of the best studied PGPR at present (Cassán and Diaz-Zorita, 2016). K. variicola UC4115 was isolated from the rhizosphere of tomato plant, after a long-term reduce tillage soil management, based on its nitrogen-fixation ability. Strains K. variicola UC4115 and A. brasilense Sp7 were screened in vitro for PGP activities related to (i) nutrition, including phosphate (P) solubilization and siderophores synthesis, (ii) the production of growth hormones like IAA, and (iii) antagonism against fungal pathogen such as Sclerotinia sclerotiorum. Later, whole genome sequencing and annotation was performed to identify target genes that contribute to the beneficial interaction between bacteria and plants. Finally, an in vivo-greenhouse experiment on tomato seedlings from BBCH (Biologische Bundesanstalt, Bundessortenamt, and CHemical industry) 11–12 to BBCH > 14 was performed in both organic and conventional management, accomplished with three inoculations (control, K. variicola UC4115, A. brasilense Sp7) at seed, root and seed and root level.
Materials and Methods
Isolation of PGPR From Rhizosphere Soil and Cultural Conditions
PGP rhizobacteria strain UC4115 was isolated from the rhizosphere of Solanum lycopersicum L., cultivated with conservation agriculture practices (i.e., reduced tillage and cover crops), in a commercial field situated in Gabbioneta-Binanuova (45°12'03.0” N; 10°12'27.8” E), Cremona, Po Valley (Northern Italy), following the method described by Guerrieri et al. (2020). Briefly, to isolate the rhizosphere bacteria, root samples were shaken vigorously to remove not tightly adhering soil. The root system was washed with sterile physiological water added with Tween 80 (0.01% v/v), and the mixture was incubated at 25°C for 90 min with shaking at 180 rpm. The resulting suspensions were serially diluted (10−3) and 0.1 mL aliquots were used to grow cultures in N-free semi-solid media, NFb (New Fabio Pedrosa), and incubated 4–7 d at 30°C until the growth of a veil-like pellicle near the surface of the culture medium, and the color change of the medium from green-blueish to blue. Subculturing the isolate three times on LB agar plates (Luria-Bertani) (Oxoid, Basingstoke, UK) resulted in pure colonies of rhizosphere bacteria.
Azospirillum brasilense Sp7 (DSMZ 1690) was provided by the German Type Culture Collection (DSMZ, Braunschweig, Germany). Bacterial strain was inoculated in LB Broth and incubated at 30°C for 48 h, with shaking at 180 rpm.
Taxonomic Identification of Bacterial Isolate UC4115
According to Guerrieri et al. (2020), the extracted DNA was amplified with 16S rRNA, using the universal primers P1 (5′-GCGGCGTGCCTAATACATGC-3′) and P6 (5′-CTACGGCTACCTTGTTACGA-3′) (Di Cello and Fani, 1996). Sanger sequencing of PCR products was carried out at GATC Biotech (Ebersberg, Germany). The obtained 16S ribosomal DNA sequences were compared with others in the GenBank database, through the NCBI-BLAST server, at https://blast.ncbi.nlm.nih.gov/Blast.cgi.
In vitro Assessment of PGP Traits
The PGP traits of K. variicola UC4115 and A. brasilense Sp7 were assessed following the methods described by Guerrieri et al. (2020). Briefly, for qualitative estimation of tri-calcium phosphate solubilization, the strains were spot inoculated on GY/Tricalcium phosphate medium (Ambrosini and Passaglia, 2017), and incubated at 30°C. After 7 days of incubation plates were observed for development of a clear halo zone around the colony, the halo's diameter was evaluated according to Ambrosini and Passaglia (2017). The phytohormone IAA production was estimated using the Salkowski reagent (12 g of FeCl3 per L in 7.9 M H2SO4) (Glickmann and Dessaux, 1995). Bacterial isolates were inoculated in LB medium supplemented with the precursor DL-Tryptophan (0.01%) and as well in LB medium without DL-Tryptophan, incubated at 30°C for 72 h, with shaking at 180 rpm. After incubation, the cultures were centrifuged at 4°C for 10 min (6,000 rpm). Equals volume of supernatant and Salkowski reagent were mixed and incubated in the dark for 30 min, then assessed for color change. Development of reddish color indicated the presence of IAA. Optical density was taken at 540 nm by using UV/visible spectrophotometer. Standard curve of IAA was used to measure the concentration of IAA produced. The strains were quantitatively assessed for siderophores production using CAS (Chrome Azurol Sulphonate) reagent (Schwyn and Neilands, 1987). Supernatant of each bacterial culture was added in separate wells of microplate followed by the addition of 100 μL CAS reagent. After 20 min optical density was taken at 620 nm using microplate reader. Siderophore production by K. variicola UC4115 and A. brasilense Sp7 was measured in percent siderophore unit (psu) which was calculated according to the following formula: [(Ar-As)/Ar] ×100 = % siderophore units. Where, Ar = absorbance of reference (CAS solution and uninoculated broth), and As = absorbance of sample (CAS solution and cell-free supernatant of sample). The two strains were also screened for antifungal activities against Sclerotinia sclerotiorum (DSM 1946) using dual culture assay (Dikin et al., 2006), on potato dextrose agar (PDA) (Oxoid, Basingstoke, UK). The antagonistic activity was observed by measuring the size of the growth inhibition zone and the percentage of growth inhibition (PGI) was calculated using the formula: [(KR–R1)/KR] ×100 = % growth inhibition. Where, KR represents the colony diameter of the pathogen in the control plate, and R1 represents the colony diameter in the treated plate.
Whole Genome Sequencing and Analysis
Genome DNA Extraction and Sequencing
Genomic DNA of K. variicola UC4115 was extracted from exponential phase LB broth culture using the E.Z.N.A Bacterial DNA kit according to the manufacturer's instruction (Omega Bio-tek, Georgia, USA). Genomic DNA was sequenced at Fasteris (Geneve, Switzerland) using an Illumina MiSeq operating with V3 chemistry in 300X2 bp paired-reads. Basecalling was performed with MiSeq Control Software 2.6.2.1, RTA 1.18.54.0, and bcl2fastq2 v2.17.1.14.
Genome Assembly and Annotation
Genome assembly was performed using PATRIC Unicycler v0.4.8 assembly pipeline (Wick et al., 2017). Genome annotation was performed both with RAST server (Aziz et al., 2008) and PATRIC RASTtk-enabled Genome Annotation Service (Brettin et al., 2015). The gene functions were further analyzed by BLASTP using Kyoto Encyclopedia of Genes Genomes (KEGG) database. The presence of plasmids was assessed by PlasmidFinder 1.3 (Carattoli et al., 2014). Phylogenetic tree was build using the codon trees pipeline in PATRIC which uses the amino acids and nucleotides sequences from PATRIC's global families (PGFams), that cross the genus boundary (Davis et al., 2016).
The genome sequence of A. brasilense Sp7 was downloaded from NCBI-Genomes database at https://www.ncbi.nlm.nih.gov/genome/. As for K. variicola UC4115, genome annotation was performed both with RAST server and PATRIC RASTtk-enabled Genome Annotation Service.
Analysis of Hemolysin Phenotype
To evaluate the hemolytic properties, K. variicola UC4115 was aerobically cultured on Blood Agar plates (Sigma-Aldrich, Germany) containing 7% (w/v) defibrinated horse blood (EO Labs, Burnhouse, Scotland) at 30°C, overnight. Staphylococcus aureus ATCC 6538 was used as a positive control for hemolysis. The development of a clear or greenish zone around the colonies is indicative of the presence of β- or α-hemolysis (Patrone et al., 2020).
Greenhouse Experiments
Inoculum's Preparation
Bacterial cultures of K. variicola UC4115 and A. brasilense Sp7 were inoculated in LB broth and incubated at 28–30°C with shaking at 180 rpm. After 14 h, cultures broths were centrifuged at 6,000 rpm for 10 min. The pellets were re-suspended in sterile distilled water (SDW) and washed thrice. The washed bacterial pellets were then reconstituted with SDW to obtain a turbid solution, whose optical density at 600 nm was adjusted to obtain a final density of 1 × 107 CFU mL−1. These solutions were used for greenhouse inoculations.
Seedling Assays
The effects of bacterial inoculation were observed over two separated greenhouse experiments: the first one using tomato variety HEINZ 3402 in a conventional commercial peat and the second one using variety HEINZ 1301 in an organic commercial peat. Details about composition of media are reported in the Supplementary Table 1. For each experiment, 280 tomato seeds were sown in a polystyrene multicell flats (280 inverted pyramid cells, 13 cm3 volume), filled with the specific commercial soil, and covered with a layer of vermiculite. Seedlings were grown under typical greenhouse growing conditions (temperatures of 18°C at night and 24°C at day, 80% relative humidity). Seedlings were irrigated daily and no fertilizer was used. Both greenhouse experiments were designed as split plot, with six replicates. Main factor was bacterial inoculation (hereafter, inoculation), with three levels: (i) K. variicola UC4115, (ii) A. brasilense Sp7 (DSMZ 1690), plus (ii) a negative control with un-inoculated seedlings. Secondary factor was the type of application (hereafter, application), with three levels: application (i) at seed level, (ii) at root level, and at (iii) both seed and root (seed plus root) level.
For seed application, tomato seeds were wetted with 10 mL of bacterial inoculum, prepared as previously described, immediately after sowing, while for root application 10 mL of bacterial suspension was applied as soil drench on the rhizosphere of 3 week-old tomato seedlings. In the third application tested, bacterial inoculation of both seeds and roots were performed as described above. In the negative control thesis, the same volume (10 mL) of sterile distilled water was used. For application at seed level, replicates were taken off from six different cells with inoculated and controls seedlings at weeks 3, 4, and 5 after sowing, corresponding to the BBCH phenological stages 11–12, 12–13, and 13–14. For application at root and at seed plus root levels, samplings from six different cells with inoculated and controls seedling were performed at weeks 4, 5, and 6 after sowing, corresponding to BBCH 12–13, BBCH 13–14, BBCH > 14. After cutting off the aerial part of the seedlings, rhizosphere soil was removed with water added with Tween 80 (0.01% v/v), in order to facilitate the separation of roots from soil. Dry weights of separated roots and shoots were weighted after heating at 105°C, overnight.
Root Characterizations
Soil samples were stored at 4°C until root separation and analysis were carried out. Determination of Root Length Density (RLD, cm cm−3) and root diameter were performed with the software winRHIZO Reg 2012. The Diameter Class Length (DCL, mm cm−3) was calculated for very fine (≤ 0.075 mm), fine (0.075–0.2 mm), medium (0.2-1.0 mm) and coarse (>1 mm) diameters, as adapted from Fiorini et al. (2018).
Statistical Analyses
Data on the evolution over time of tomato seedlings above-ground and below-ground biomass in both experiments (i.e., conventional and organic) were subjected to analysis of variance (ANOVA) with a mixed-effect model using the “nlme” package of RStudio3.3.3 (Pinheiro et al., 2017). Phenological stages (BBCH) were included in the model as fixed factor while block effect was considered as random. Repeated measures were used to assess the effects of inoculation with K. variicola UC4115, A. brasilense Sp7 and negative control at seed level, root level, and seed plus root level over time. Since BBCH stages for application at seed level were different from those for application at root and at seed plus root level, analyses were performed separately for each type of application.
Data on the final effect of inoculation on root length density (RLD), and diameter class length (DCL) for very fine (Ø ≤ 0.075 mm), fine (Ø = 0.075–0.2 mm), medium (Ø = 0.2–1.0 mm), and coarse (Ø ≥ 1.0 mm) diameters were statistically analyzed with split-plot ANOVA.
When normality of variances was not confirmed using the Sharpiro-Wilk test, data were log transformed before analysis. Mean values were separated with Tukey honestly significant difference (HSD) test (α = 0.05), using the “Estimated Marginal Means, aka Least-Squares Means” package, version 1.2.4 (Lenth et al., 2018).
Results
Phenotypic Features of the Two Tested PGPR
For both strains K. variicola UC4115 and A. brasilense Sp7 the growth of a veil-like pellicle near the surface of the culture medium NFb was observed, moreover, the color change of the medium from green-blueish to blue was another indicator for the bacterial growth (Table 1). The capability of both strains to grow on nitrogen-free medium indicated their putative activity as diazotrophic bacteria.
Table 1.
Plant growth promotion properties of K. variicola UC4115 and A. brasilense Sp7.
| PGP property | K. variicola UC4115 | A. brasilense Sp7 |
|---|---|---|
| Growth on N-free agar medium | Growth and change color observed | Growth and change color observed |
| Phosphate solubilization | Level 2 | – |
| IAA production (w/Try; w/o Try) | 68.32 μg/ml; 3.16 μg/ml | 3.06 μg/ml; 3.18 μg/ml |
| Siderophore production | 41.50 psu | 1.90 psu |
| Biocontrol activity against Sclerotinia sclerotiorum | 33.33%. | 41.54% |
The isolate K. variicola UC4115 was able to solubilize the phosphate of the GY/Tricalcium phosphate medium by producing clear zone around the colonies after 7 days of incubation. Considering the halo's diameter bigger than 0 cm up to 1 cm, K. variicola strain UC4115 was classified as Level 2 phosphate solubilizer (Table 1). A. brasilense Sp7 strain was not able to grow on GY/Tricalcium phosphate medium (Table 1), so its capability to solubilize phosphate was not evaluated in vitro.
To screen for indole-3-acetic acid (IAA) production the Salkowski reagent was used, which gave different degree of red to the solution according to the different levels of IAA produced. The concentration of IAA produced by K. variicola strain UC4115 showed variation in presence of L-Tryptophan (68.32 μg mL−1) and without the IAA precursor tryptophan (3.16 μg mL−1) (Table 1). The concentration of IAA produced by strain A. brasilense Sp7 was very low both in presence (3.06 μg mL−1) and in absence (3.18 μg mL−1) of the IAA precursor tryptophan (Table 1).
The quantitative estimation of siderophore production was carried out using CAS reagent. Optical density revealed that the concentration of siderophore produced by K. variicola UC4115 measured 41.50 psu, while measured 1.90 psu for A. brasilense Sp7 (Table 1).
Antifungal activity, of both strains K. variicola UC4115 and A. brasilense Sp7, was tested against Sclerotinia sclerotiorum (DSM 1946) using the dual plate technique, showing a percentage of growth inhibition (PGI) corresponding to 33.33% for K. variicola UC4115, and 41.54% for A. brasilense Sp7 (Table 1).
As a safety measure, the activity of hemolysins for K. variicola UC4115 were tested. However, neither β-hemolytic nor α-hemolytic phenotypes were detected on blood agar plates. In contrast, the positive control S. aureus ATCC 6538 exhibited the expected β-hemolysin activity (data not shown).
Genomic Features of the Two Tested PGPR
The general genomic properties of K. variicola UC4115 and A. brasilense Sp7 are presented in Table 2. The presence of one plasmid was verified using PlasmidFinder (v1.3), the plasmid was similar to plasmid pCAV1099-114 of K. oxytoca strain CAV1099, with 96% identity. We established a phylogenetic tree based on the PGFams, global proteins families that cross the genus boundary, with 100 conserved genes, zero deletions and zero duplications allowed. The tree supported the 16S results, confirming that the strain UC4115 is most closely related to K. variicola (Figure 1).
Table 2.
General features of K. variicola UC4115 and A. brasilense Sp7 genomes.
| Feature | K. variicola UC4115 | A. brasiliense Sp7 |
|---|---|---|
| Size (bp) | 5,539,38 bp | 7,100,241 bp |
| G + C content (%) | 57.34 | 68.41 |
| Number of CDSs | 5,378 | 6,667 |
| tRNA | 77 | 74 |
| rRNA | 4 | 9 |
| Plasmid | 1 | 5 |
| Coverage | 309x | 105x |
Figure 1.
Phylogeny of K. variicola UC4115 based on the analysis of PGFams.
Genome annotation was performed with PATRIC RASTtk-enabled Genome Annotation Service and further confirmed with RAST server. Relating to nitrogen fixation ability K. variicola UC4115 genome harbored nifJHDKTYENXUSVWZMFLABQ genes together with the rnf ABCDEG operon (Table 3). The genes nifMFLJ, and the rnf cluster, were absent from A. brasilense Sp7. However, A. brasilense Sp7 genome contained the fixABCXJ genes (Table 3).
Table 3.
Genes annotation of K. variicola UC4115 and A. brasilense Sp7 genomes related to nitrogen fixation, phosphate solubilization, IAA production, siderophores synthesis, and biocontrol activity.
| Trait | PGfams ID | Gene annotation | KO/Gene_ID | KEGG_GENE_NAME | UC 4115 | Sp7 |
|---|---|---|---|---|---|---|
| Nitrogenase complex | PGF_00025954 | Nitrogenase iron protein | K02588 | nifH | * | * |
| PGF_00025951 | Nitrogenase molybdenum-iron protein alpha chain | K02586 | nifD | * | * | |
| PGF_00025953 | Nitrogenase molybdenum-iron protein beta chain | K02591 | nifK | * | * | |
| PGF_00120348 | Putative nitrogen fixation protein FixT | K02593 | nifT | * | * | |
| PGF_00025669 | NifY protein | – | nifY | * | * | |
| PGF_00025964 | Nitrogenase MoFe cofactor biosynthesis protein NifE | K02587 | nifE | * | * | |
| PGF_00945843 | Nitrogenase iron-molybdenum-cofactor biosynthesis protein NifN | K02592 | nifN | * | * | |
| PGF_00025961/ PGF_00025962 |
Nitrogen fixation protein NifX | K02596 | nifX | * | * | |
| PGF_00420050 | Cysteine desulfurase NifS | K04487 | nifS | * | * | |
| PGF_00072326 | Homocitrate synthase | K02594 | nifV | * | * | |
| PGF_01958698 | Putative NifW protein | K02595 | nifW | * | * | |
| PGF_00025671 | Putative NifZ protein | K02597 | nifZ | * | * | |
| PGF_01971790 | Nitrogen fixation protein NifM | K03769 | nifM | * | ||
| PGF_00071382 | Flavodoxin FldA | K03839 | nifF | * | ||
| PGF_00029105 | Nitrogen fixation negative regulator NifL/PAS-PAC protein | K23916 | nifL | * | ||
| PGF_03973235 | Nif-specific regulatory protein | K02584 | nifA | * | * | |
| PGF_06674514 | Nitrogenase cofactor biosynthesis protein NifB | K02585 | nifB | * | * | |
| PGF_00025976 | Nitrogen fixation protein NifQ | K15790 | nifQ | * | * | |
| PGF_00046007 | Putative pyruvate:ferredoxin (flavodoxin) oxidoreductase | K03737 | nifJ | * | ||
| PGF_00015530 | Iron-sulfur cluster assembly scaffold protein | K04488 | nifU | * | * | |
| PGF_00401722 | Nitrogenase-associated protein NifO | – | nifO | * | * | |
| Nitrogenase transport | PGF_00424131 | Na(+)-translocating NADH-quinone reductase subunit E | K03617 | rnfA | * | |
| PGF_10542033 | Electron transport complex protein RnfB; Required for nitrogen fixation | K03616 | rnfB | * | ||
| PGF_04400591 | Electron transport complex protein rnfC | K03615 | rnfC | * | ||
| PGF_01678333 | Electron transport complex protein RnfD; Required for nitrogen fixation | K03614 | rnfD | * | ||
| PGF_01484108 | NADH-ubiquinone oxidoreductase | K03613 | rnfE | * | ||
| PGF_00424141 | Electron transport complex protein RnfG | K03612 | rnfG | * | ||
| Electron transport | PGF_05015473 | Ferredoxin-like protein FixX/4Fe-4S ferredoxin | K03855 | FixX | * | |
| PGF_03136394 | Electron transfer flavoprotein-quinone oxidoreductase FixC | K00313 | FixC | * | ||
| PGF_03134445 | Electron transfer flavoprotein, alpha subunit FixB | K03522 | FixB | * | ||
| PGF_03098759 | Electron transfer flavoprotein, beta subunit FixA | K03521 | FixA | * | ||
| PGF_00849787 | Two-component system response regulator | K14987 | FixJ | * | ||
| Gluconic acid | PGF_04577966 | Quinoprotein glucose dehydrogenase/glucose dehydrogenase, PQQ-dependent | K00117 | gcd | * | |
| PGF_01393943 | Pyrroloquinoline-quinone synthase C | K06137 | pqqC | * | * | |
| PGF_01084777 | Coenzyme PQQ synthesis protein F | – | pqqF | * | ||
| PGF_00418484 | Coenzyme PQQ synthesis protein B | K06136 | pqqB | * | * | |
| PGF_03579782 | Coenzyme PQQ synthesis protein D | K06138 | pqqD | * | ||
| PGF_00418486 | Coenzyme PQQ synthesis protein E | K06139 | pqqE | * | * | |
| Phosphonate transporter | PGF_12684827 | Phosphonate ABC transporter, permease protein | K02042 | phnE1 | * | |
| PGF_00033841 | Phosphonate ABC transporter, permease protein | K02042 | phnE2 | * | ||
| PGF_00033852 | Phosphate-binding protein of phosphonate ABC transporter | K02044 | phnD | * | ||
| PGF_00033832 | Phosphonate transport system ATP-binding protein | K02041 | PhnC | * | ||
| Phosphate transporter | PGF_07668761 | Phosphate transport system substrate-binding protein | K02040 | Pst S | * | * |
| PGF_01072302 | Phosphate transport system permease protein | K02038 | Pst A | * | * | |
| PGF_02405545 | Phosphate transport system permease protein | K02037 | Pst C | * | * | |
| PGF_06213055 | Phosphate transport system ATP-binding protein | K02036 | Pst B | * | * | |
| Indole-3-acetic acid (IAA) biosynthesis | PGF_05599542 | Indole-3-pyruvate decarboxylase | K04103 | ipdC | * | * |
| PGF_00049805 | Amidase | K01426 | – | * | ||
| PGF_07597988 | Aldehyde dehydrogenase (NAD+) (EC 1.2.1.3) | aldh | * | * | ||
| PGF_00418275 | Nitrile hydratase, alpha subunit | K01721 | nthA | * | * | |
| PGF_00418276 | Nitrile hydratase, beta subunit | K20807 | nthB | * | * | |
| PGF_03811905/ PGF_02254418 |
Histidinol-phosphate aminotransferase | K00817 | hisC | * | * | |
| Siderophore production | PGF_00015658/ PGF_00015659 |
Isochorismate/ Apo-aryl carrier protein | K01252 | entB, dhbB, vibB, mxcF | * | * |
| PGF_00025850/ PGF_07637567 |
2,3-dihydroxybenzoate-AMP ligase | K02363 | entE, dhbE, vibE, mxcE | * | * | |
| PGF_05075091 | Enterobactin synthetase component F | K02364 | entF | * | * | |
| PGF_00424602 | Enterobactin exporter | K08225 | entS | * | ||
| PGF_00023831/ PGF_08225224 |
2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | K00216 | entA | * | * | |
| PGF_00015696 | Isochorismate synthase | K02361 | entC | * | ||
| PGF_00422373 | 4′-phosphopantetheinyl transferase EntD | K02362 | entD | * | ||
| PGF_00037591 | Proofreading thioesterase in enterobactin biosynthesis | K24147 | entH | * | ||
| PGF_07721642 | MbtH-like protein | K05375 | MbtH | * | ||
| PGF_00004447 | Ferric enterobactin-binding periplasmic protein | K23185 | FepB | * | ||
| PGF_00004441 | Ferric enterobactin transport system permease protein | K23186 | FepD | * | ||
| PGF_00004444 | Ferric enterobactin transport system permease protein | K23187 | FepG | * | ||
| PGF_00004439 | Ferric enterobactin transport ATP-binding protein | K23188 | FepC | * | ||
| PGF_00424600/ PGF_08225224 |
Enterobactin esterase | K07214 | Fes | * | * | |
| PGF_00057226 | TonB-dependent receptor; Outer membrane receptor for ferric enterobactin and colicins B, D | K19611 | FepA | * | * | |
| PGF_00052044 | Alternative sigma factor | – | PvdS | * | ||
| PGF_00045754 | PvdE, pyoverdine ABC export system, fused ATPase and permease components | K06160 | PvdE | * | ||
| 4-hydroxybenzoate Production | PGF_00417843 | Chorismate-pyruvate lyase | K03181 | ubiC | * | |
| GABA | PGF_07204877 | Succinate-semialdehyde dehydrogenase | K00135 | gabD | * | * |
| PGF_04337880 | 4-aminobutyrate aminotransferase | K07250 | gabT | * | * | |
| Phenazine biosynthesis | PGF_10329977 | Phenazine biosynthesis | - | phzF | * | * |
The symbol * indicates that the gene is present in the genome.
Regarding phosphorus solubilization ability, K. variicola UC4115 genome harbored gcd and pqqBCDEF genes. Moreover, phosphate (PstABCS) and phosphonate (PhnCDE1E2) transporter system genes were detected (Table 3). The genes gcd, pqqFD, and phn cluster were absent from A. brasilense Sp7 (Table 3).
Gene prediction identified genes specifically associated with IAA production. Indeed, both strains K. variicola UC4115 and A. brasilense Sp7 carried ipdC, aldh, and nthAB genes, the gene amidase was absent from A. brasilense Sp7 (Table 3).
Relating to the synthesis of siderophores K. variicola UC4115 carried entABCDEFGHS genes together with several genes for siderophores receptors such as FepABCDG. While, A. brasilense Sp7 harbored PvdSE genes (Table 3).
Regarding to biocontrol activities UbiC, gabDT, and phzF genes were detected in K. variicola UC4115 genome. All the defense genes detected in K. variicola UC4151 strain was also detected in A. brasilense Sp7 strain, with the exception of UbiC (Table 3).
Seedling Assays
Evolution Over Time of Tomato Seedling Biomass Under Conventional Management
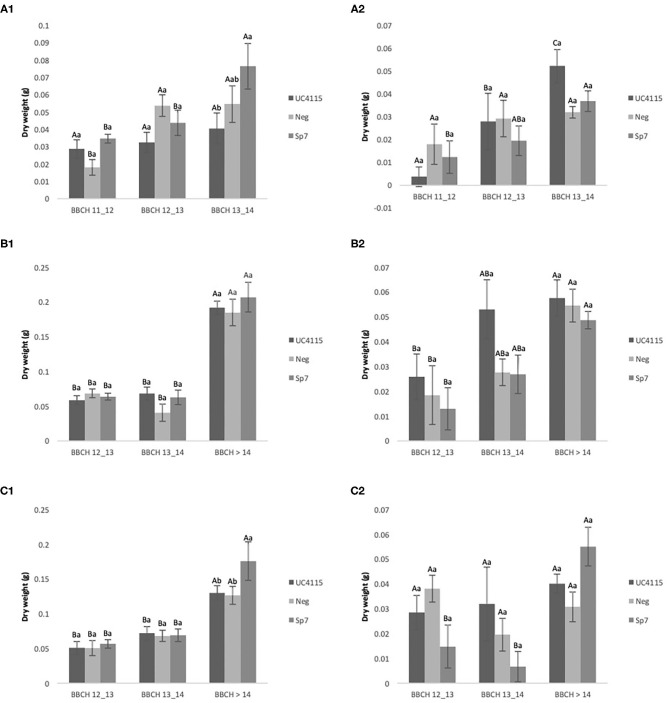
After application at seed level, negative control and A. brasilense Sp7 increased above-ground biomass of tomato seedlings passing from BBCH 11–12 to BBCH 13–14. Conversely, K. variicola UC4115 did not lead to any above-ground biomass variation between the two phenological stages. The inoculation hierarchy at BBCH 13–14 was A. brasilense Sp7 ≥ negative control ≥ K. variicola UC4115 (Figure 2A1). Below-ground biomass of tomato seedlings was increased from BBCH 11–12 to BBCH 13–14 by K. variicola UC4115, while not by A. brasilense Sp7 and negative control. This turns into higher biomass in K. variicola UC4115 than in A. brasilense Sp7, while the negative control did not differ from both the former and the latter (Figure 2A2).
Figure 2.
Conventional experiment. Evolution over time of tomato plant above-ground biomass (A1,B1,C1) and below-ground biomass (A2,B2,C2) after inoculation with UC4115, Sp7, and negative control at seed level (A1,A2), root level (B1,B2), and seed plus root level (C1,C2). Mean values ± standard deviation. Capital letters indicated differences among physiological state (BBCH) of tomato plant within the same inoculation; lowercase letters indicate differences among different inoculations within the same physiological state.
After application at root level, the above-ground biomass of tomato seedling was affected by all inoculations: K. variicola UC4115, A. brasilense Sp7 and negative control increased above-ground biomass from BBCH 12-13 to BBCH >14, and no difference between inoculations occurred at each phenological stage (Figure 2B1). A similar pattern was observed for the below-ground biomass of tomato seedlings, which increased with increasing BBCH stage (Figure 2B2). Also, after the application at seed and root level, K. variicola UC4115, A. brasilense Sp7 and negative control increased above-ground biomass of tomato seedling passing from BBCH 12–13 to BBCH > 14. Differences between inoculations occurred only at to BBCH >14, when A. brasilense Sp7 had higher above-ground biomass than K. variicola UC4115 and negative control (Figure 2C1). As regards below-ground biomass of tomato seedlings, a few differences were found between and within each BBCH stage: only A. brasilense Sp7 increased values from BBCH 12–13 to BBCH > 14 (Figure 2C2).
On overall, the results indicated that the inoculation at seed- level and the co-inoculation at seed plus root level were the more effective applications, while the root application had shown less evidence in terms of dry root and dry shoot biomass. Further, these data indicated that the effects, on root and shoot dry mass, are more evident when the inoculations were made at the latest stages of growth after sowing.
Evolution Over Time of Tomato Seedling Biomass Under Organic Management
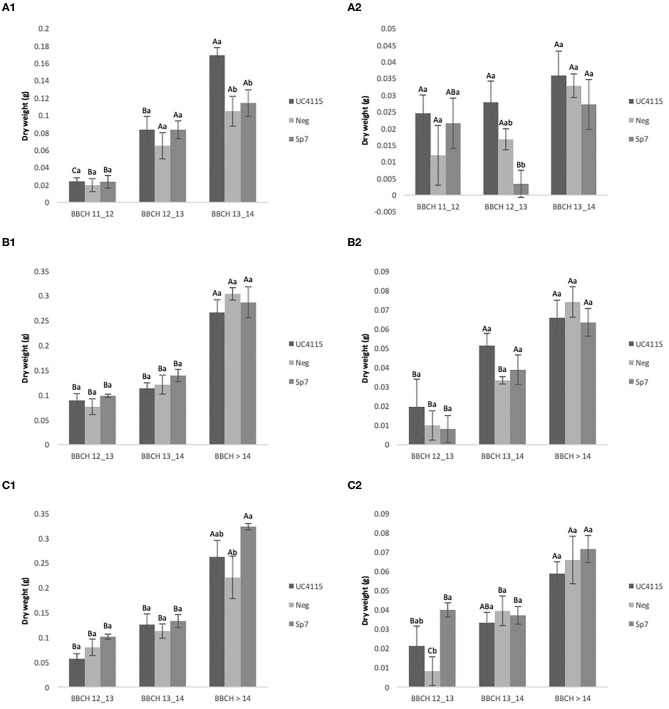
After application at seed level, all inoculations increased above-ground biomass of tomato seedling passing from BBCH 11–12 to BBCH 13–14. Differences between inoculations occurred at BBCH 13–14, when K. variicola UC4115 had higher biomass than A. brasilense Sp7 and negative control, where the biomass increment was 62% when compared to negative control (Figure 3A1). Trends of below-ground biomass of tomato seedlings did not show a specific pattern and differences between inoculations were found only at BBCH 13–14, when A. brasilense Sp7 was lower than K. variicola UC4115 and negative control (Figure 3A2).
Figure 3.
Organic experiment. Evolution over time of tomato plant above-ground biomass (A1,B1,B2) and below-ground biomass (A2,B2,C2) after inoculation with UC4115, Sp7, and negative control at seed level (A1,A2), root level (B1,B2) and seed plus root level (C1,C2). Mean values ± standard deviation. Capital letters indicated differences among physiological state (BBCH) of tomato plant within the same inoculation; lowercase letters indicate differences among different inoculations within the same physiological state.
After application at root level, the above-ground and below-ground biomasses of tomato seedling were not significantly affected by inoculations at any BBCH stage (Figures 3B1,B2). K. variicola UC4115, A. brasilense Sp7 and negative control increased both biomass fractions from BBCH 12–13 to BBCH > 14. Similarly, above-ground biomass of tomato seedling was increased from BBCH 12–13 to BBCH > 14 by all inoculations, after application at seed and root level. However, A. brasilense Sp7 induced higher biomass than Neg at BBCH > 14, while K. variicola UC4115 did not differ from both the former and the latter (Figure 3C1). Also, below-ground biomass increased passing from BBCH 12–13 to BBCH > 14 in all inoculations. Nevertheless, differences here were found at BBCH 12–13, when negative control was lower that A. brasilense Sp7 (Figure 3C2).
Even under organic management the inoculation at seed level and the co-inoculation at seed plus root level, were more effective than the root application, that had shown no evidence in terms of dry root and shoot mass.
Root Length Density (RLD) and Diameter Class Length (DCL) of Tomato Seedlings at the End of Organic Management
Root length density of tomato seedlings at the end of the experiment was significantly affected by inoculation, while not by application. In detail, K. variicola UC4115 had on average higher RLD than A. brasilense Sp7, while negative control did not differ from the former and the latter. The interaction I × T was also significant and K. variicola UC4115 with application at root level had higher RLD than A. brasilense Sp7 with application at seed- and root level. All the others were in the between (Table 4).
Table 4.
Root length density (RLD), and diameter class length (DCL) for very fine (Ø = 0.00–0.075 mm), fine (Ø = 0.075–0.2 mm), medium (Ø = 0.2–1.0 mm), and coarse (Ø = > 1.0 mm) diameters for organic experiment, as affected by inoculation (I) and application (A).
| DCL (cm cm–3) | |||||||
|---|---|---|---|---|---|---|---|
| Source of variation | Inoculation | RLD (cm cm–3) | Ø = 0.00–0.075 mm | Ø = 0.075–0.2 mm | Ø = 0.2–1.0 mm | Ø = > 1.0 mm | |
| Inoculation | UC4115 | 300.105a | 23.047 | 81.870 | 192.533a | 2.655ab | |
| Neg | 284.640ab | 21.475 | 81.851 | 177.983ab | 3.331a | ||
| Sp7 | 261.074b | 20.759 | 75.530 | 162.712b | 2.074b | ||
| P-value | <0.01 | – | – | <0.01 | <0.05 | ||
| DCL (cm cm–3) | |||||||
| Source of variation | Application | RLD (cm cm–3) | Ø = 0.00–0.075 mm | Ø = 0.075–0.2 mm | Ø = 0.2–1.0 mm | Ø = > 1.0 mm | |
| Application | S | 285.964 | 22.824 | 78.106 | 182.047 | 2.987 | |
| R | 292.869 | 21.923 | 86.415 | 181.684 | 2.847 | ||
| SR | 266.986 | 20.535 | 74.730 | 169.495 | 2.226 | ||
| P-value | – | – | – | – | – | ||
| DCL (cm cm–3) | |||||||
| Source of variation | Inoculation | Application | RLD (cm cm–3) | Ø = 0.00–0.075 mm | Ø = 0.075–0.2 mm | Ø = 0.2–1.0 mm | Ø = > 1.0 mm |
| I × A | UC4115 | S | 308.273ab | 22.464 | 78.140 | 204.094a | 3.574 |
| R | 322.774a | 24.495 | 91.952 | 203.985a | 2.342 | ||
| SR | 269.268ab | 22.182 | 75.516 | 169.520ab | 2.050 | ||
| Neg | S | 280.265ab | 22.976 | 83.701 | 170.836ab | 2.752 | |
| R | 288.240ab | 21.612 | 88.628 | 173.933ab | 4.068 | ||
| SR | 285.414ab | 19.839 | 73.224 | 189.179ab | 3.172 | ||
| Sp7 | S | 269.354ab | 23.033 | 72.476 | 171.212ab | 2.634 | |
| R | 267.592ab | 19.661 | 78.664 | 167.136ab | 2.131 | ||
| SR | 246.276b | 19.583 | 75.450 | 149.787b | 1.456 | ||
| P-value | <0.05 | – | – | <0.05 | – | ||
Values followed by the same letter in the each column are not statistically significant according to Tukey's HSD test.
Diameter class length for very fine and fine roots was never affected by inoculation or application, as well as by the interaction I × T. Significant differences were found in DCL for medium (with Inoculation and I × T as sources of variation) and coarse roots (with Inoculation as source of variation): (i) Inoculation as source of variation showed higher DCL for medium roots with K. variicola UC4115 than with A. brasilense Sp7, while I × T showed higher DCL for medium roots under K. variicola UC4115 with application at seed-, and at root level than under A. brasilense Sp7 with seed plus root level; (ii) coarse roots had higher DCL on average under negative control than under A. brasilense Sp7, while K. variicola was in the between (Table 4).
Root Length Density (RLD) and Diameter Class Length (DCL) of Tomato Seedlings at the End of Conventional Management
Inoculation significantly affected RLD of tomato seedlings at the end of the experiment. In detail, K. variicola UC4115 and A. brasilense Sp7 had on average higher RLD than negative control. Also, the interaction I × T was found to be significant: RLD under K. variicola UC4115 with application at root-, and at seed plus root level, and under A. brasilense Sp7 with application at root level was higher than that under negative control with application at the root level. All the other combinations were in the between (Table 5).
Table 5.
Root length density (RLD), and diameter class length (DCL) for very fine (Ø = 0.00–0.075 mm), fine (Ø = 0.075–0.2 mm), medium (Ø = 0.2–1.0 mm), and coarse (Ø = > 1.0 mm) diameters for conventional experiment, as affected by inoculation (I) and application (A).
| DCL (cm cm–3) | |||||||
|---|---|---|---|---|---|---|---|
| Source of variation | Inoculation | RLD (cm cm–3) | Ø = 0.00–0.075 mm | Ø = 0.075–0.2 mm | Ø = 0.2–1.0 mm | Ø = > 1.0 mm | |
| Inoculation | UC4115 | 140.844a | 26.150 | 32.763 | 81.217 | 0.714 | |
| Neg | 107.470b | 19.881 | 27.110 | 60.063 | 0.417 | ||
| Sp7 | 134.758a | 22.980 | 32.320 | 78.700 | 0.759 | ||
| P-value | <0.01 | – | – | – | – | ||
| DCL (cm cm–3) | |||||||
| Source of variation | Application | RLD (cm cm–3) | Ø = 0.00–0.075 mm | Ø = 0.075–0.2 mm | Ø = 0.2–1.0 mm | Ø = > 1.0 mm | |
| Application | S | 120.514 | 18.973 | 29.787 | 696.694 | 0.794 | |
| R | 128.489 | 24.865 | 31.078 | 732.041 | 0.633 | ||
| SR | 134.070 | 25.173 | 31.328 | 771.058 | 0.463 | ||
| P-value | – | – | – | – | – | ||
| DCL (cm cm–3) | |||||||
| Source of variation | Inoculation | Application | RLD (cm cm–3) | Ø = 0.00–0.075 mm | Ø = 0.075–0.2 mm | Ø = 0.2–1.0 mm | Ø = > 1.0 mm |
| I × A | UC4115 | S | 114.228ab | 14.089 | 28.808ab | 71.331ab | 0.000b |
| R | 151.681a | 34.438 | 34.509a | 81.894a | 0.840ab | ||
| SR | 156.624a | 29.924 | 34.973a | 90.425a | 1.301a | ||
| Neg | S | 125.262ab | 18.933 | 32.325ab | 72.831ab | 1.174a | |
| R | 77.398b | 20.219 | 18.699b | 38.442b | 0.037b | ||
| SR | 119.751ab | 20.491 | 30.305ab | 68.915ab | 0.040b | ||
| Sp7 | S | 122.052ab | 23.898 | 32.100ab | 64.846ab | 1.208a | |
| R | 156.387a | 19.936 | 36.153a | 99.276a | 1.022a | ||
| SR | 125.836ab | 25.105 | 28.707ab | 71.977ab | 0.046ab | ||
| P-value | <0.01 | – | <0.05 | <0.01 | <0.001 | ||
Values followed by the same letter in the each column are not statistically significant according to Tukey's HSD test.
The interaction I × T significantly affected also the response of DCL for fine, medium, and coarse roots: in any case K. variicola UC4115 with application at seed- and root level had the highest DCL, while negative control with application at the root level the lowest (Table 5).
Discussion
Phenotypic and Genomic Features of the Two Tested PGPR
In the current study K. variicola UC4115 was isolated from the rhizosphere and rhizoplane soil of tomato plants, from a field with a long history of reduced tillage plus cover crops management. The isolate was screened for biocontrol and PGP activities according to Guerrieri et al. (2020), showing high growth-promoting and defense abilities. We performed a functional annotation of genes contributing to the five plant-beneficial functions previously tested in-vitro. Furthermore, the in-vitro characterization, the whole genome sequencing and annotation, performed with K. variicola UC4115, were compared with the well-known PGPR Azospirillum brasilense strain Sp7 (GenBank: GCA_001315015.1). Azospirillum sp. is able to colonize hundreds of plant species and significantly improves their growth, development and productivity (Cassán and Diaz-Zorita, 2016). In detail strain A. brasilense Sp7 is well-characterized free-nitrogen fixing bacteria, particularly known for its anchoring capabilities at root surface (Ramirez-Mata et al., 2018). Hence, this global comparative study allowed to increment the knowledge about the newly isolated strain K. variicola UC4115 and to observe how different genera of PGPR affect differently the plant host.
According to the functional annotation, K. variicola UC4115 strain contained a number of genes corroborating the in-vitro results. Both strains K. variicola UC4115 and A. brasilense Sp7 were able to grown on nitrogen-free medium, this indicates their putative activity as diazotrophic bacteria. Indeed, according to literature both species are among the major nitrogen-fixing bacteria detected in root system (Bhattacharjee et al., 2008). Nitrogenase is the enzyme that catalyzes the conversion of atmospheric N2 to a bio-accessible form of nitrogen, and it consists of Fe-protein encoded by nifH (component I) and MoFe-protein encoded by nifDK (component II). Furthermore, the gene cluster from Klebsiella genome has been a model system for studying nitrogen fixation and consists in a total of 20 genes, nifJHDKTYENXUSVWZMFLABQ (Rubio and Ludden, 2008). The K. variicola UC4115 genome contains all the above nif genes together with the rnf ABCDEG operon, which encodes a putative membrane-bound complex related to electron transport to nitrogenase (Jeong and Jouanneau, 2000). Even if, genes nifMFLJ, and the rnf cluster, were absent from A. brasilense Sp7, this strain genome contained the fix genes (fixABCXJ) that are essential for nitrogen fixation but do not have an homologous counterpart in Klebsiella genus (Fischer, 1994). Fix genes were especially detected in rhizobia such as Bradyrhizobium japonicum and Azorhizobium caulinodans (Tsoy et al., 2016). In detail, it has been discovered that the fixABCX gene products probably replace the missing NifJ and NifF electron transfer proteins in rhizobia, that operate in Klebsiella. Moreover, it was previously suggested that nif gene number might vary according to the physiology of a bacterium. Indeed, alternatively, unidentified proteins might replace the missing Nif products (Masson-Boivin et al., 2009).
The capability of K. variicola UC4115 to solubilize the insoluble source of phosphate, contained in the GY/Tricalcium medium, was further confirmed through the identification of genes associated with phosphate solubilization. Gluconic acid (GA) is an organic acid recognized as one of the major responsible for the solubilization of mineral phosphate in most bacteria. GA biosynthesis is catalyzed by glucose-1-dehydrogenase (GDH) and its co-factor pyrrolo-quinolone quinine (PQQ) (Ramachandran et al., 2006). The K. variicola UC4115 genome possesses several genes related to GA biosynthesis and its co-factor genes, including pqqBCDEF (Wagh et al., 2014), while the gene pqqA is lacking. Researchers (Liu et al., 2016), during the study of the genome of the PGPR Klebsiella sp. D5A, have already reported the lacking of this specific gene. The enzyme encoded by pqqA would appear not essential for the biosynthesis of PQQ (Toyama and Lidstrom, 1998). Furthermore, the uptake of the inorganic P in K. variicola UC4115 may be promoted by the high affinity with the phosphate transporter system, PstABCS, and the phosphonate transporter system, PhnCDE1E2 (Liu et al., 2016; Shariati et al., 2017), indeed various studies have indicate that phosphonates are another rich source of soil P (Oliverio et al., 2020). The genes gcd, pqqFD and phn cluster were absent from A. brasilense Sp7. Moreover, A. brasilense Sp7 strain was not able to grow on GY/Tricalcium phosphate medium, so its capability to solubilize phosphate was not evaluated in vitro. Usually the pqqBCDE are very conservative genes shared by different type of PGPR, indeed, PQQ is a co-factor implicated in several cellular process (Bruto et al., 2014). However, the incapability of Azospirillum sp. to solubilize P since they lack the pqq genes was already evaluated (Vikram et al., 2007).
According to literature the IAA-producing potential of species K. variicola is another dominant feature as PGPR, together with nitrogen fixation (Kim et al., 2017). In Trp-dependent IAA biosynthesis, four pathways have been postulated: (i) the indole-3-acetamide (IAM) pathways; (ii) the indole-3-pyruvate (IPA) pathways; (iii) the tryptamine (TAM) pathways; and (iv) the indole-3-acetonitrile (IAN) pathway (Spaepen and Vanderleyden, 2011). Here, two proposed pathways, the IAN and IPA pathways, are identified in the genome of K. variicola UC4115. Indeed, the isolate carries the nitrile hydratase (nthAB) and the amidase genes, which contribute, respectively, to the conversion of indole-3-acetonitrile (IAN) in indole-3-acetamide (IAM) and finally in IAA, and the ipdC gene, the key enzyme for the indole-3-pyruvate (IPA) decarboxylation (Spaepen and Vanderleyden, 2011). These two pathways have been also detected in the genome of the PGPR Klebsiella sp. D5A (Liu et al., 2016). The presence of ipdC gene in the A. brasilense Sp7 genome confirm the presence of the IPA pathways. Nonetheless, the concentration of IAA produced by strain A. brasilense Sp7 was very low both in presence (3.06 μg mL−1) and in absence (3.18 μg mL−1) of the IAA precursor tryptophan. However, in 1992 Bar and Okon, found out that indole-3-acetic acid production of A. brasilense Sp7 depended on culture age and amount of tryptophan supplement (Bar and Okon, 1992). During our study we only tested one concentration of tryptophan at 0.01%, to analyze the effective capability of producing IAA, several concentrations should be compared.
Relating to the siderophores production, K. variicola UC4115 carried genes entABCDEFGH which catalyze the conversion of chorismate, an intermediate of aromatic amino acid synthesis, into the enterobactin (Hubrich et al., 2021), and the gene entS, which is responsible for the transport of the siderophore. Furthermore, K. variicola strain UC4115 encodes several genes for siderophores receptors (FepBCDG), including TonB-dependent receptor outer-membrane receptor (FepA) (Shariati et al., 2017). The genes entS, entCDH, and the Fep cluster were absent from A. brasilense Sp7. This seemed to confirm the extremely low value (1.90 psu), of siderophore produced by the strain A. brasilense Sp7. However, according to literature A. brasilense strain Sp7 seems to be able to secrete a catecholtype siderophore called spirilobactin, that can chelate and transport iron (Alahari et al., 2006; Tortora et al., 2011). Moreover, our genomic study did not detect the presence of genes involved in pyoverdine synthesis (Pvd), one of the major class of siderophores (Liu et al., 2016), in K. variicola UC4115 genome, by contrast PvdSE were detected in A. brasilense Sp7 genome.
Biocontrol activities are important mechanisms by which plant growth promoting bacteria suppress plant pathogens (Shen et al., 2013). It has been stated that PGPR may produce molecules able to suppress plant pathogenic microbes or are responsible for pest and disease inhibition. The common compound produced by PGPR and major studied so far are 4-hydroxybenzoate and γ-aminobutyric acid (GABA). UbiC is the involved in 4-hydroxybenzoate synthesis, while gabD and gabT genes contribute to GABA synthesis (Gupta et al., 2014). Despite of the low biocontrol activity against S. sclerotiorum, K. variicola UC4115 genome contains all the above genes. Moreover, also the gene phzF, involved in the synthesis of the antibiotic phenazine (Gupta et al., 2014) was detected. The higher biocontrol activity of A. brasilense, against S. sclerotiorum, could be explain by the production of the siderophore pyoverdine, indeed the biocontrol activity of these molecules seems to be involved in the biocontrol activity of Pseudomonas fluorescens (Shen et al., 2013).
Seedling Assays
Under organic management, K. variicola UC4115 showed the greatest performance at seed level. The effects were always evident at the latest stages of growth after sowing. The increment of dry shoot biomass extended both phenotypic and genomic characterization, underlying its capability as nitrogen-fixing rhizobacteria, phosphate solubilizers, and siderophores producer, that are those abilities that confer a major impact on the above-ground biomass. Moreover, this result could also be affected by the greatest influence, of K. variicola UC4115, on length and diameters of the roots. This is certainty due to the significant amount of IAA produced by the strain confirmed by the genome analysis and in-vitro screening. Auxin can regulate the size of the root apical meristems (Aloni et al., 2006). In detail, IAA helps in the production of longer roots with increased number of root hairs and root laterals which are involved in nutrient uptake (Mohite, 2013). The IAA effects seemed more evident after application at root- and seed-level. Data at root-level seemed to show a negative tendency between biomass values and RLD and DCL values. Presumably the greater RLD was due to the increased root thickness, and RLD did not translate into high root and shoot biomass (Nada and Abogadallah, 2018).
The capability of A. brasilense Sp7 to increment the dry shoot biomass under conventional management, at both seed and seed-root application, reflected its high value trait as nitrogen-fixing rhizobacteria and underlined the genomic characterization. Indeed, several studies stated the importance of the role of fixABC cluster and some of the nif genes in the A. brasilense Sp7 as high nitrogen-fixing rhizobacteria (Galimand et al., 1989). Furthermore, Jankiewicz, in 2006, observed how magnesium ions present in the bacterial growth medium of Pseudomonas, improved the synthesis of siderophore pyoverdine (Jankiewicz, 2006), by assumption, the Mg within the conventional peat, could have stimulated the Pvd genes detected in A. brasilense Sp7 genome. Moreover, the higher performance of A. brasilense Sp7 compared to K. variicola UC4115, could be explained by the presence of fertilizers in the commercial peat, hence, the positive effects of association between strains of A. brasilense inoculations and N rates, are frequently reported in literature (Marini et al., 2015). In addition, undoubtedly, interactions between plant genotype and bacteria, are one of the most contributing factors to the complexity of responses to inoculation (Rodriguez et al., 2019). The capability of A. brasilense Sp7 to affect the RLD and DCL after root application, confirmed the genomic evaluation performed and the presence of IPA pathway, despite of the in-vitro results. This was in accord with the scientific literature about the IAA-producing potential of strain A. brasilense Sp7 (Castro-Guerrero et al., 2012). For both K. variicola UC4115 and A. brasilense Sp7 we observed a negative tendency between biomass and RLD and DCL parameters.
Conclusion
In conclusion, in this study, we present a comprehensive genomic and greenhouse evaluation of PGPRs, using the multifunctional K. variicola UC4115 as a case study. The screening in-vitro results were confirmed and supported by the genome analysis, demonstrating that K. variicola UC4115 acted as PGPR, indeed the strain contained many of the signature genes that are functionally linked to the plant growth promotion traits. Comparative in-vitro screening and genomic analyses with the well-known PGPR A. brasilense Sp7 revealed different accumulation of PBFC genes, and potential PGPR features not detected through the in-vitro assay. Comparative greenhouse assays on tomato seedling allowed to observed how these differences affect the plant growth. Here, we report for the first time a holistic comparative discussion between phenotypic analysis, genomic characterization and plant study. The comparison between functional annotation and in-vivo results, of two different PGPR, allowed us to better understand their different effects on tomato plant, also extending the results from in-vitro assay. Indeed, this study highlight the importance of genome analysis as predictor of PGPR behavior on plant growth. We believe that the holistic comparative discussion, could enhance the overview of the issue about PGPR mode of actions on their eukaryotic host, cultured under greenhouse condition. Further studies are firmly necessary to establish the usefulness of molecular mechanism in less controlled environment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI SRA BioProject PRJNA699239: UC4115.
Author Contributions
MG, EP, and MT: conceptualization. EP, MT, PC, and VT: methodology. MG, EF, and AF: formal analysis and writing— original draft. MG, EF, and EP: investigation. MG, AF, EF, EP, and VT: writing—review and editing. EP and MT: supervision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the Consultancy Agency Expedia s.r.l. (Piacenza, Italy) for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.660620/full#supplementary-material
References
- Alahari A., Tripathi A. K., Le Rudulier D. (2006). Cloning and characterization of a fur homologue from Azospirillum brasilense Sp7. Curr. Microbiol. 52, 123–127. 10.1007/s00284-005-0204-x [DOI] [PubMed] [Google Scholar]
- Aloni R., Aloni E., Langhans M., Ullrich C. I. (2006). Role of cytokinin and auxin in shaping root architecture : regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 97, 883–893. 10.1093/aob/mcl027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini A., Passaglia L. M. P. (2017). Plant growth-promoting bacteria (pgpb): isolation and screening of PGP activities. Curr. Protoc. plant Biol. 2, 190–209. 10.1002/pb.20054 [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 1–15. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar T., Okon Y. (1992). Induction of indole-3-acetic acid synthesis and possible toxicity of tryptophan in Azospirillum brasilense Sp 7. Symbiosis 13, 191–198. [Google Scholar]
- Barrios-Camacho H., Aguilar-Vera A., Beltran-Rojel M., Aguilar-Vera E., Duran-Bedolla J., Rodriguez-Medina N., et al. (2019). Molecular epidemiology of Klebsiella variicola obtained from different sources. Sci. Rep. 9, 1–10. 10.1038/s41598-019-46998-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee R. B., Singh A., Mukhopadhyay S. N. (2008). Use of nitrogen-fixing bacteria as biofertiliser for non-legumes: prospects and challenges. Appl. Microbiol. Biotechnol. 80, 199–209. 10.1007/s00253-008-1567-2 [DOI] [PubMed] [Google Scholar]
- Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S., Olsen G. J., et al. (2015). RASTtk : a modular and extensible implementation of the RAST algorithm for annotating batches of genomes. Sci. Rep. 5:8365. 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruto M., Prigent-Combaret C., Muller D., Moënne-Loccoz Y. (2014). Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 4, 1–10. 10.1038/srep06261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., Garciá-Fernández A., Larsen M. V., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassán F., Diaz-Zorita M. (2016). Azospirillum sp. in current agriculture: from the laboratory to the field. Soil Biol. Biochem. 103, 117–130. 10.1016/j.soilbio.2016.08.020 [DOI] [Google Scholar]
- Castro-Guerrero J., Romero A., Aguilar J. J., Xiqui M. L., Sandoval J. O., Baca B. E. (2012). The hisC1 gene, encoding aromatic amino acid aminotransferase-1 in Azospirillum brasilense Sp7, expressed in wheat. Plant Soil 356, 139–150. 10.1007/s11104-011-1009-2 [DOI] [Google Scholar]
- Chen M., Li Y., Li S., Tang L., Zheng J., An Q. (2016). Genomic identification of nitrogen-fixing Klebsiella variicola, K. pneumoniae and K. quasipneumoniae. J. Basic Microbiol. 56, 78–84. 10.1002/jobm.201500415 [DOI] [PubMed] [Google Scholar]
- Davis J. J., Gerdes S., Olsen G. J., Olson R., Pusch G. D., Shukla M., et al. (2016). PATtyFams: protein families for the microbial genomes in the PATRIC database. Front. Microbiol. 7:118. 10.3389/fmicb.2016.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cello F., Fani R. (1996). A molecular strategy for the study of natural bacterial communities by PCR-based techniques. Minerva Biotecnol. 8, 126–134. [Google Scholar]
- Dikin A., Sijam K., Kadir J., Semanz I. A. (2006). Antagonistic bacteria against Schizophyllum commune FR. In Peninsular Malaysia. Biotropia 13, 111–121. 10.11598/btb.2006.13.2.221 [DOI] [Google Scholar]
- Fiorini A., Boselli R., Amaducci S., Tabaglio V. (2018). Effects of no-till on root architecture and root-soil interactions in a three-year crop rotation. Eur. J. Agron. 99, 156–166. 10.1016/j.eja.2018.07.009 [DOI] [Google Scholar]
- Fischer H. M. (1994). Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58, 352–386. 10.1128/MR.58.3.352-386.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M., Perroud B., Delorme F., Paquelin A., Vieille C., Bozouklian H., et al. (1989). Identification of DNA regions homologous to nitrogen fixation genes nifE, nifUS and fixABC in Azospirillum brasilense Sp7. J. Gen. Microbiol. 135, 1047–1059. 10.1099/00221287-135-5-1047 [DOI] [PubMed] [Google Scholar]
- Glickmann E., Dessaux Y. (1995). A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61, 793–796. 10.1128/AEM.61.2.793-796.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrieri M. C., Fanfoni E., Fiorini A., Trevisan M., Puglisi E. (2020). Isolation and screening of extracellular PGPR from the rhizosphere of tomato plants after long-term reduced tillage and cover crops. Plants 9:668. 10.3390/plants9050668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Gopal M., Thomas G. V., Manikandan V., Gajewski J., Thomas G., et al. (2014). Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS ONE 9:e104259. 10.1371/journal.pone.0104259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubrich F., Andexer J. N., Andexer J., Mu M., Mu M. (2021). Chorismate- and isochorismate converting enzymes: versatile catalysts acting on an important metabolic node. R. Soc. Chem. 57, 2441–2463. 10.1039/D0CC08078K [DOI] [PubMed] [Google Scholar]
- Jankiewicz U. (2006). Synthesis of siderophores by soil bacteria of the genus Pseudomonas under various culture conditions. Acta Sci. Pol. Agric. 5, 33–44. [Google Scholar]
- Jeong H. S., Jouanneau Y. (2000). Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J. Bacteriol. 182, 1208–1214. 10.1128/JB.182.5.1208-1214.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. Y., Shahzad R., Kang S. M., Seo C. W., Park Y. G., Park H. J., et al. (2017). IAA-producing Klebsiella variicola AY13 reprograms soybean growth during flooding stress. J. Crop Sci. Biotechnol. 20, 235–242. 10.1007/s12892-017-0041-0 [DOI] [Google Scholar]
- Lenth R., Singmann H., Love J., Buerkner P., Herve M. (2018). Package ‘emmeans'. Estimated marginal measn, aka least-squares means. R Packag. version 1.15–15. [Google Scholar]
- Liu W., Hou J., Wang Q., Ding L., Luo Y. (2014). Isolation and characterization of plant growth-promoting rhizobacteria and their effects on phytoremediation of petroleum-contaminated saline-alkali soil. Chemosphere 117, 303–308. 10.1016/j.chemosphere.2014.07.026 [DOI] [PubMed] [Google Scholar]
- Liu W., Wang Q., Hou J., Tu C., Luo Y., Christie P. (2016). Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6, 1–10. 10.1038/srep26710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini D., Guimarães V. F., Dartora J., do Lana M. C., Pinto Júnior A.S. (2015). Growth and yield of corn hybrids in response to association with Azospirillum brasilense and nitrogen fertilization. Rev. Ceres 62, 117–123. 10.1590/0034-737X201562010015 [DOI] [Google Scholar]
- Martínez-Viveros O., Jorquera M. A., Crowley D. E., Gajardo G., Mora M. L. (2010). Mechanisms and practical considerations involved in plant growth promotion by Rhizobacteria. J. Soil Sci. Plant Nutr. 10, 293–319. 10.4067/S0718-95162010000100006 [DOI] [Google Scholar]
- Masson-Boivin C., Giraud E., Perret X., Batut J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. 10.1016/j.tim.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Mohite B. (2013). Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 13, 638–649. 10.4067/S0718-95162013005000051 [DOI] [Google Scholar]
- Nada R. M., Abogadallah G. M. (2018). Root-favoured biomass allocation improves growth and yield of field-gown rice (Oryza sativa L.) plants only when the shoot sink is expandable. Acta Physiol. Plant 40:123. 10.1007/s11738-018-2697-5 [DOI] [Google Scholar]
- Oleńska E., Małek W., Wójcik M., Swiecicka I., Thijs S., Vangronsveld J. (2020). Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: a methodical review. Sci. Total Environ. 743:140682. 10.1016/j.scitotenv.2020.140682 [DOI] [PubMed] [Google Scholar]
- Oliverio A. M., Bissett A., McGuire K., Saltonstall K., Turner B. L., Fierera N. (2020). The role of phosphorus limitation in shaping soil bacterial communities and their metabolic capabilities. Ecol. Evol. Sci. 11, 1–16. 10.1128/mBio.01718-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olowe O. M., Akanmu A. O., Asemoloye M. D. (2020). Exploration of microbial stimulants for induction of systemic resistance in plant disease management. Ann. Appl. Biol. 117, 282–293. 10.1111/aab.12631 [DOI] [Google Scholar]
- Patrone V., Al-Surrayai T., Romaniello F., Fontana A., Milani G., Sagheddu V., et al. (2020). Integrated phenotypic-genotypic analysis of candidate probiotic Weissella cibaria strains isolated from dairy cows in Kuwait. Probiot. Antimicrob. Proteins. 10.1007/s12602-020-09715-x. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J., Bates D., DebRoy S., Sarkar D., Heisterkam S., Van Willige B., et al. (2017). nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-131. Available online at: https://CRAN.R-project.org/package=nlme
- Podile A. R., Kishore G. K. (2006). Plant growth-promoting rhizobacteria, in Plant-Associated Bacteria (Dordrecht: Springer; ), 195–230. [Google Scholar]
- Ramachandran S., Fontanille P., Pandey A., Larroche C. (2006). Gluconic acid: properties, applications and microbial production. Food Technol. Biotechnol. 44, 185–195. [Google Scholar]
- Ramirez-Mata A., Pacheco M. R., Moreno S. J., Xiqui-Vazquez M. L., Baca B. E. (2018). Versatile use of Azospirillum brasilense strains tagged with egfp and mCherry genes for the visualization of biofilms associated with wheat roots. Microbiol. Res. 215, 155–163. 10.1016/j.micres.2018.07.007 [DOI] [PubMed] [Google Scholar]
- Rodriguez P. A., Rothballer M., Chowdhury S. P., Nussbaumer T., Gutjahr C., Falter-Braun P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant 12, 804–821. 10.1016/j.molp.2019.05.006 [DOI] [PubMed] [Google Scholar]
- Rubio L. M., Ludden P. W. (2008). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111. 10.1146/annurev.micro.62.081307.162737 [DOI] [PubMed] [Google Scholar]
- Ruzzi M., Aroca R. (2015). Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 196, 124–134. 10.1016/j.scienta.2015.08.042 [DOI] [Google Scholar]
- Schwyn B., Neilands J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- Sekhar A. C., Thomas P. (2015). Isolation and identification of shoot-tip associated endophytic bacteria from Banana cv. Grand Naine and testing for antagonistic activity against Fusarium oxysporum f. sp. cubense. Am. J. Plant Sci. 6, 943–954. 10.4236/ajps.2015.67101 [DOI] [Google Scholar]
- Shariati V. J., Malboobi M. A., Tabrizi Z., Tavakol E., Owilia P., Safari M. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 7, 1–12. 10.1038/s41598-017-15820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Hu H., Peng H., Wang W., Zhang X. (2013). Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics 14:271. 10.1186/1471-2164-14-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V. K., Singh A. K., Singh P. P., Kumar A. (2018). Interaction of plant growth promoting bacteria with tomato under abiotic stress: a review. Agric. Ecosyst. Environ. 267, 129–140. 10.1016/j.agee.2018.08.020 [DOI] [Google Scholar]
- Spaepen S., Vanderleyden J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. 10.1101/cshperspect.a001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V. B., Rajamohan G. (2020). Comparative genome analysis and characterization of a MDR Klebsiella variicola. Genomics 112, 3179–3190. 10.1016/j.ygeno.2020.06.004 [DOI] [PubMed] [Google Scholar]
- Sunera A., Saqib S., Uddin S., Zaman W., Ullah F., Ayaz A., et al. (2020). Characterization and phytostimulatory activity of bacteria isolated from tomato (Lycopersicon esculentum Mill.) rhizosphere. Microb. Pathog. 140:103966. 10.1016/j.micpath.2020.103966 [DOI] [PubMed] [Google Scholar]
- Tortora M. L., Díaz-Ricci J. C., Pedraza R. O. (2011). Azospirillum brasilense siderophores with antifungal activity against Colletotrichum acutatum. Arch. Microbiol. 193, 275–286. 10.1007/s00203-010-0672-7 [DOI] [PubMed] [Google Scholar]
- Toyama H., Lidstrom M. E. (1998). pqqA is not required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1. Microbiology 144, 183–191. 10.1099/00221287-144-1-183 [DOI] [PubMed] [Google Scholar]
- Tsoy O. V., Ravcheev D. A., Cuklina J., Gelfand M. S. (2016). Nitrogen fixation and molecular oxygen: comparative genomic reconstruction of transcription regulation in Alphaproteobacteria. Front. Microbiol. 7:1343. 10.3389/fmicb.2016.01343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J., Desbrosses G., Bouffaud M. L., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4, 1–19. 10.3389/fpls.2013.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejan P., Abdullah R., Khadiran T., Ismail S., Nasrulhaq Boyce A. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability-a review. Molecules 21:573. 10.3390/molecules21050573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram A., Alagawadi A. R., Krishnaraj P. U., Mahesh Kumar K. S. (2007). Transconjugation studies in Azospirillum sp. negative to mineral phosphate solubilization. World J. Microbiol. Biotechnol. 23, 1333–1337. 10.1007/s11274-007-9365-z [DOI] [Google Scholar]
- Wagh J., Shah S., Bhandari P., Archana G., Kumar G. N. (2014). Heterologous expression of pyrroloquinoline quinone (pqq) gene cluster confers mineral phosphate solubilization ability to Herbaspirillum seropedicae Z67. Appl. Microbiol. Biotechnol. 98, 5117–5129. 10.1007/s00253-014-5610-1 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, 1–22. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Yang K. (2020). Biological function of Klebsiella variicola and its effect on the rhizosphere soil of maize seedlings. PeerJ 8, 1–24. 10.7717/peerj.9894 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI SRA BioProject PRJNA699239: UC4115.