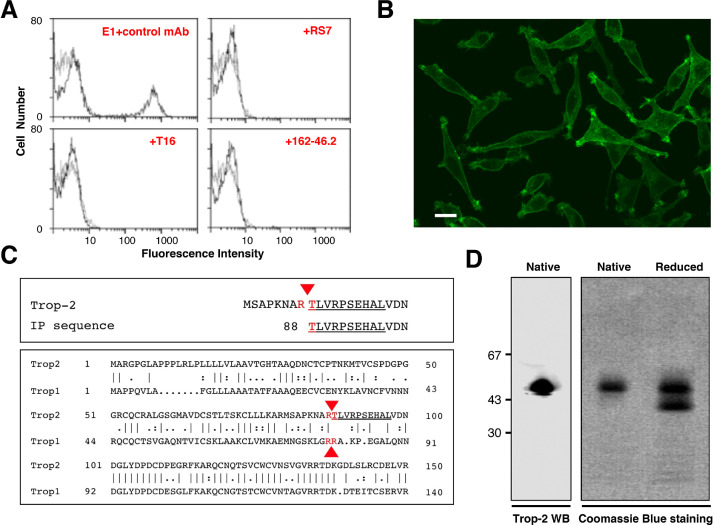
Fig. 1.
Purification, sequencing and analysis of Trop-2. (A) Reconstituted mixtures of 70% parental L cells and 30% Trop-2/L cells transfectants [34] were used for competition studies of E1 with other anti-Trop-2 mAbs. Cell mixtures were stained with the FITC-E1 mAb. Competition of E1 with other anti-Trop-2 mAbs was performed by incubating cell mixtures with 100-fold excess of the indicated ascites (red). Successful competition was revealed by the disappearance of the peak of stained cells, indicating that the E1 binding site is the same as, or is in close proximity to, that of the competing Abs. E1 was efficiently competed-out by the anti-Trop-2 162-46.2 antibody [80], the T16 mAb [13] and the RS7-3G11 mAb, from which the humanized anti-Trop-2 therapeutic IMMU132 was derived [47]. (B) Reactivity of the E1 mAb with Trop-2. Immunofluorescence microscopy analysis of FITC-E1-stained TROP2/L cell transfectants. (C, top) Alignment of the amino-acid sequence obtained by Edman degradation (underlined) with the canonical Trop-2 sequence. Red arrowhead: cleavage site. (C, bottom) Alignment of the proteolytic sites of Trop-2 and Trop-1 [21] (red arrows). The Edman degradation sequence of E1/Trop-2 is underlined. (D, left) Western blotting of Trop-2, as immunoprecipitated from ovarian cancer cells and purified by affinity chromatography over Sepharose–E1 mAb. Purified Trop-2 was run in SDS-PAGE under native (nonreducing) conditions. (D, right) Coomassie blue staining of purified Trop-2 protein run in SDS-PAGE gradient gel under native or reducing conditions. (Colored version of figure is available online.)