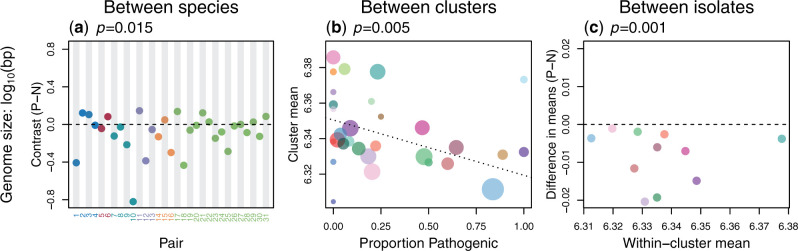
Fig. 2.
Pathogenicity is associated with smaller genomes. (a) Each point represents a phylogenetically independent species pair, with numbers and colors from figure 1a. Standardized contrast values are ordered such that negative values imply smaller genomes in the pathogenic species. P-value is from a permutation test of the null hypothesis of no difference in the mean contrast value (as indicated by the dotted line). (b) Each point represents a cluster of Streptococcus suis isolates, with colors from figure 1b, and sizes indicating the number of isolates. A sample-size weighted regression shows that clusters containing a larger proportion of pathogenic isolates have a smaller average genome size. (c) Each point shows the difference in mean genome size between disease and carriage isolates in a cluster of S. suis isolates, such that negative values imply smaller genomes in the pathogens. Each point corresponds to a “mixed cluster” (containing multiple disease and carriage isolates), with colors and numbers matching figure 1b. The P-value is from a permutation test as in (a).