Summary
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection leading to coronavirus disease 2019 (COVID-19) usually results in respiratory disease, but extrapulmonary manifestations are of major clinical interest. Intestinal symptoms of COVID-19 are present in a significant number of patients, and include nausea, diarrhea, and viral RNA shedding in feces. Human induced pluripotent stem cell-derived intestinal organoids (HIOs) represent an inexhaustible cellular resource that could serve as a valuable tool to study SARS-CoV-2 as well as other enteric viruses that infect the intestinal epithelium. Here, we report that SARS-CoV-2 productively infects both proximally and distally patterned HIOs, leading to the release of infectious viral particles while stimulating a robust transcriptomic response, including a significant upregulation of interferon-related genes that appeared to be conserved across multiple epithelial cell types. These findings illuminate a potential inflammatory epithelial-specific signature that may contribute to both the multisystemic nature of COVID-19 as well as its highly variable clinical presentation.
Keywords: human induced pluripotent stem cells, intestinal and colonic organoids, SARS-CoV-2, extrapulmonary COVID-19, host response, intestinal epithelial cells
Highlights
-
•
Human iPSC-derived intestinal organoids are susceptible to productive SARS-CoV-2 infection
-
•
SARS-CoV-2 infection is accompanied by ultrastructural changes in HIO morphology
-
•
RNA-seq revealed a common inflammatory signature across multiple epithelial cell types
-
•
HIOs are a viable tool to model host response to infectious pathogens in the GI tract
In this article, Gustavo Mostoslavsky, Elke Mühlberger, and colleagues model the host response to intestinal SARS-CoV-2 infection using iPSC-derived regionally patterned intestinal organoids. Specifically, they highlight productive infection of mesenchyme-free intestinal and colonic organoids and demonstrate that they recapitulate intrinsic inflammatory transcriptomic responses to infection that have been shown in other epithelial cell types.
Introduction
Over the past two decades, multiple zoonotic coronaviruses have caused outbreaks of human respiratory diseases. Most notably, the spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Zhou et al., 2020b) and its subsequent manifestation as coronavirus disease 2019 (COVID-19) has caused a global pandemic that has led to over 100 million documented cases and over two million deaths globally as of January 2021 (Dong et al., 2020). Although the primary manifestations of COVID-19 involve the respiratory system, there is increasing evidence that gastrointestinal (GI) manifestations of SARS-CoV-2 infection may play a critical role in both disease severity and transmission (Pan et al., 2020; Wang et al., 2020; Xiao et al., 2020). This is exemplified by the first confirmed COVID-19 patient in the United States, who presented with GI symptoms and had both fecal and respiratory specimens test positive for SARS-CoV-2 RNA (Holshue et al., 2020). It has also been shown that up to 53% of hospitalized patients tested positive for SARS-CoV-2 RNA in stool, and 23% of these patients continued to test positive for SARS-CoV-2 RNA in stool despite having negative respiratory samples (Xiao et al., 2020). Various biopsy samples from COVID-19 patients revealed interstitial edema in the lamina propria of the stomach, duodenum, and rectum, with numerous infiltrating lymphocytes, in addition to the presence of SARS-CoV-2 nucleocapsid protein by immunofluorescence microscopy at all three sites (Xiao et al., 2020). A multicohort study on the role of GI symptoms in hospitalized COVID-19 patients found that GI involvement was associated with lower mortality, reduced disease severity, reduced levels of circulating inflammatory cytokines, and less intestinal inflammation (Livanos et al., 2020). However, earlier studies out of Hubei Province showed that patients with GI symptoms had worse outcomes and longer hospitalizations (Pan et al., 2020) and yet another recent study found no association between GI involvement and the requirement for mechanical ventilation or death (Elmunzer et al., 2020), indicative that further investigation into the role of GI infection in COVID-19 disease progression is needed.
The SARS-CoV-2 entry receptor, angiotensin-converting enzyme 2 (ACE2) (Hoffmann et al., 2020; Zhou et al., 2020b), is expressed in the adult GI epithelium (Hamming et al., 2004; To and Lo, 2004) and has been shown to play a critical role in dietary amino acid homeostasis (Hashimoto et al., 2012). Notably, single-cell RNA sequencing (RNA-seq) has revealed differential ACE2 mRNA expression at various sites in the GI tract, with higher ACE2 mRNA levels reported in small intestinal enterocytes compared with colonic epithelium (Qi et al., 2020). In addition, TMPRSS2, a serine protease that is required for efficient SARS-CoV-2 entry in some cell types (Hoffmann et al., 2020), is highly expressed in human small intestinal and colonic epithelium (Vaarala et al., 2001; Zang et al., 2020). Potential regional-specific differences in SARS-CoV-2 infection are of major interest, particularly in the context of pre-existing regionally specific GI disease, as well as new reports of intestinal dysbiosis in patients who have recovered from COVID-19 (Yeoh et al., 2021; Zuo et al., 2020), highlighted by reduced levels of immunomodulatory commensals that correlated with increased levels of inflammatory markers up to 30 days after disease resolution. We have previously shown that human induced pluripotent stem cell (iPSC)-derived intestinal organoids (HIOs) express both ACE2 and TMPRSS2 mRNA at levels comparable with primary controls (Abo et al., 2020), making them promising target cells for SARS-CoV-2 infection in vitro, particularly in the context of modeling GI infection and clinical significance in COVID-19.
There are multiple reports that adult stem cell (ASC)-derived intestinal organoids can be productively infected by SARS-CoV-2 (Lamers et al., 2020; Stanifer et al., 2020; Zang et al., 2020; Zhou et al., 2020a). Analysis of the host response in these organoids revealed the importance of type I and III interferon signaling in controlling viral replication and spread, which has been previously shown to play a role in intestinal epithelial response to viral infection, including other coronaviruses such as SARS-CoV-1 (Mahlakoiv et al., 2012). The usefulness of these systems is hindered, however, by the limited availability of the primary tissue from which they are generated. Given the clinical and epidemiological implications of SARS-CoV-2 infection of the GI tract, human iPSC-derived proximal intestinal and colonic organoids represent a potentially powerful tool to study SARS-CoV-2 intestinal infection, since they are both amenable to genetic manipulation and infinitely expandable, making them particularly useful for high-throughput drug screening applications, as has been recently reported (Han et al., 2020). Furthermore, a reductionist, well-characterized epithelial organoid system enables the direct interrogation of epithelial-intrinsic responses to viral infection in the absence of immune or mesenchymal cells, particularly in the context of recent work detailing the significance of type I interferon and inflammation signaling in COVID-19 clinical outcomes (Bastard et al., 2020; Hadjadj et al., 2020; Zhang et al., 2020). Importantly, members of our group have recently utilized iPSC-derived type 2 alveolar cells (iAT2s) to study the epithelial-intrinsic response to SARS-CoV-2 infection in the distal lung, implicating interferon and nuclear factor κB (NF-κB) signaling as a key host response pathway (Hekman et al., 2021; Huang et al., 2020).
Here, we report the establishment of two robust iPSC-derived organoid models of GI SARS-CoV-2 infection. We recently established a novel, mesenchyme-free directed differentiation protocol that generates regionally patterned proximal (small intestinal) and colonic organoids from human iPSCs using a CDX2-eGFP reporter iPSC line (BU1CG) (Mithal et al., 2020). These iPSC-derived HIOs are devoid of mesenchymal cells, contain a variety of cell types found in the intestinal epithelium, including goblet, enteroendocrine, and Paneth cells, and were shown to be useful in modeling other diseases of the GI epithelium (Mithal et al., 2020). The ability to study SARS-CoV-2 infection in organoids that recapitulate both proximal and distal intestinal epithelium provides a unique opportunity to identify mechanisms of viral replication, spread, and host response signatures that could contribute to COVID-19 disease severity and progression.
Results
HIOs Are Permissive to SARS-CoV-2 Infection
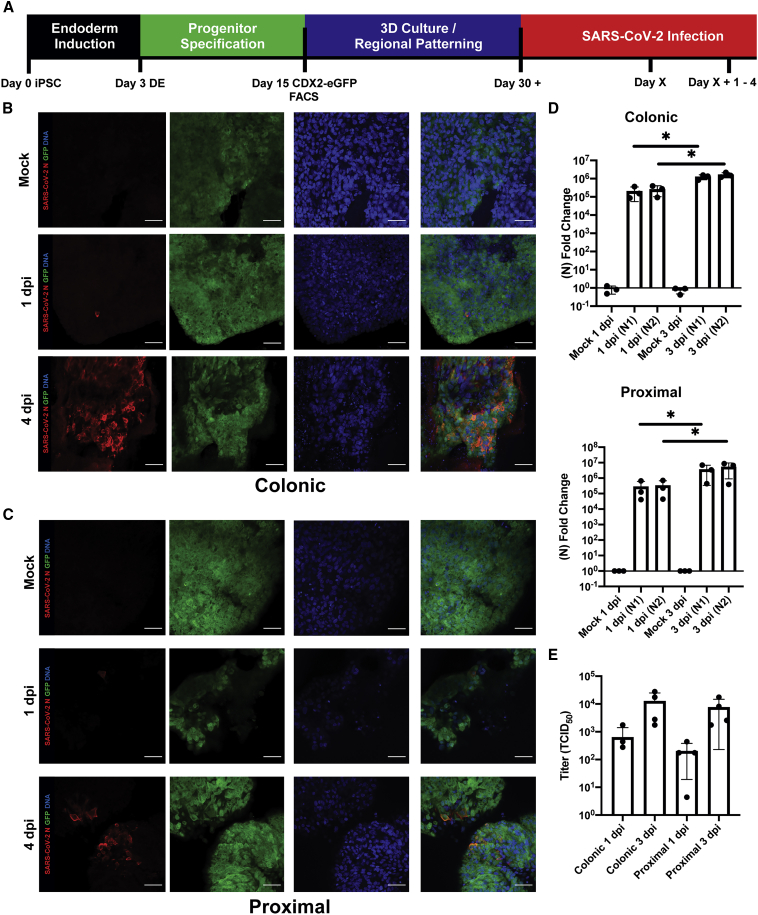
BU1CG iPSCs were differentiated into regionally patterned intestinal organoids, as previously described (Figure 1A and Mithal et al., 2020). At day 15 (Figure S1A), cells were sorted to isolate the CDX2-GFP+ population, consisting of putative intestinal epithelial progenitors (Figure 1A). Sorted cells were then cultured in 3D Matrigel droplets, in media to promote the emergence of either proximal intestinal or colonic organoids. Organoids were then infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.4. Both colonic and proximal organoids were readily infected by SARS-CoV-2, as demonstrated by the presence of the viral nucleocapsid protein (N) (Figures 1B, 1C, and S1B–S1F) and viral RNA (Figure 1D). Both viral RNA levels and the number of SARS-CoV-2-positive cells increased over time, indicative of viral replication and spread. In line with these results, viral titers in the cell supernatants were low at 1-day post infection (dpi) and increased by about 2 log at 3 dpi, indicating the production and release of infectious SARS-CoV-2 virions in the HIO (Figure 1E, n = 4 wells from separate infection experiments). The titers were comparable in proximal and colonic cells. Infection was also quantified by flow cytometry, with an average of 11.59% ± 1.75% of cells infected across both conditions at 1 dpi and an average of 20.8% ± 4.34% of cells at 4 dpi (n = 4 wells from separate infection experiments) (Figure S1G).
Figure 1.
SARS-CoV-2 Infects iPSC-Derived Human Intestinal Organoids
(A) Experimental schematic for directed differentiation of BU1CG iPSC into HIOs and SARS-CoV-2 infection.
(B) Representative confocal images of colonic BU1CG HIOs. Whole-mounted mock- and SARS-CoV-2-infected colonic HIOs at 1 and 4 dpi stained for SARS-CoV-2 N and GFP (CDX2) (scale bar,50 μm).
(C) Representative immunofluorescent confocal micrographs of proximally patterned BU1CG. Whole-mounted mock- and SARS-CoV2-infected proximal (small intestinal) HIOs at 1 and 4 dpi stained for SARS-CoV-2 N and GFP (CDX2) (scale bar,50 μm, images representative of n = 3 replicate directed differentiations).
(D) qRT-PCR for two sequences of the SARS-CoV-2 N gene in both proximal and colonic HIOs 1 and 3 dpi (2−ΔΔCT, technical duplicates normalized to mock, GAPDH, n = 3 independent directed differentiations, error bars represent the SD, statistical significance where indicated determined by unpaired Student's t test, ∗p < 0.05).
(E) Viral titers (TCID50) of culture media from 1 to 3 dpi colonic and proximal HIOs.
See also Figure S1.
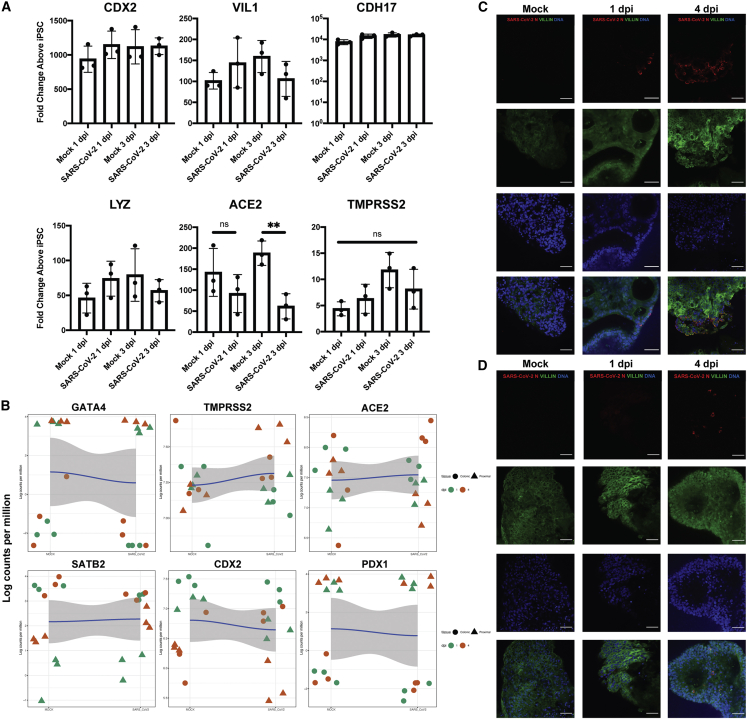
We then sought to determine whether SARS-CoV-2 infection affected HIO intestinal phenotype by performing quantitative real-time PCR (qRT-PCR) on infected colonic organoids for a variety of intestinal-specific genes. Based on the expression of intestinal-HOX gene CDX2, brush border cytoskeletal components VIL1 (villin) and CDH17, and Paneth cell marker LYZ, there were no significant differences between SARS-CoV-2 and mock-infected colonic HIOs (Figure 2A). Consistent with other reports (Huang et al., 2020), there was a significant decrease in SARS-CoV-2 entry receptor ACE2 mRNA in infected cells at 3 dpi, whereas TMPRSS2 expression was not affected by infection (Figure 2A). These data were confirmed by immunofluorescence microscopy showing robust TMPRSS2 and Villin staining in infected cells at 3 and 4 dpi (Figures S2A and S2B). Both colonic and proximal organoids retained their regional intestinal transcriptional programs, as shown by maintained expression of CDX2, colonic-specific expression of SATB2, and proximal-specific expression of GATA4 and PDX2 in both mock and infected organoids (Figure 2B). Confocal microscopy of both colonic (Figure 2C) and proximal (Figure 2D) HIOs showed maintained expression of Villin at both 1 and 4 dpi, implying that SARS-CoV-2 infection does not produce major alterations to the intestinal transcriptional program.
Figure 2.
HIOs Maintain Their Regional Identity after SARS-CoV-2 Infection
(A) qRT-PCR for phenotypic intestinal markers at 1 and 3 dpi colonic HIOs (2−ΔΔCT, technical duplicates normalized to GAPDH, n = 3 independent directed differentiation experiments, error bars represent the SD, statistical significance where indicated determined by unpaired Student's t test, ∗∗ = p < 0.01).
(B) Local regression (locally estimated scatterplot smoothing [LOESS]) plots profiling regional-specific (colonic/proximal) markers in mock (left) and infected (right) HIOs.
(C) Representative confocal immunofluorescent micrographs of infected colonic HIOs showing maintained expression of Villin, at 1 and 4 dpi.
(D) Representative confocal immunofluorescent micrographs of infected proximal HIOs showing maintained expression of Villin, at 1 and 4 dpi. (All scale bars represent 50 μm, images representative of n = 3 replicate directed differentiations).
See also Figures S1 and S2.
SARS-CoV-2 Induces Structural Changes in Infected HIOs that Support the Production of Viral Particles
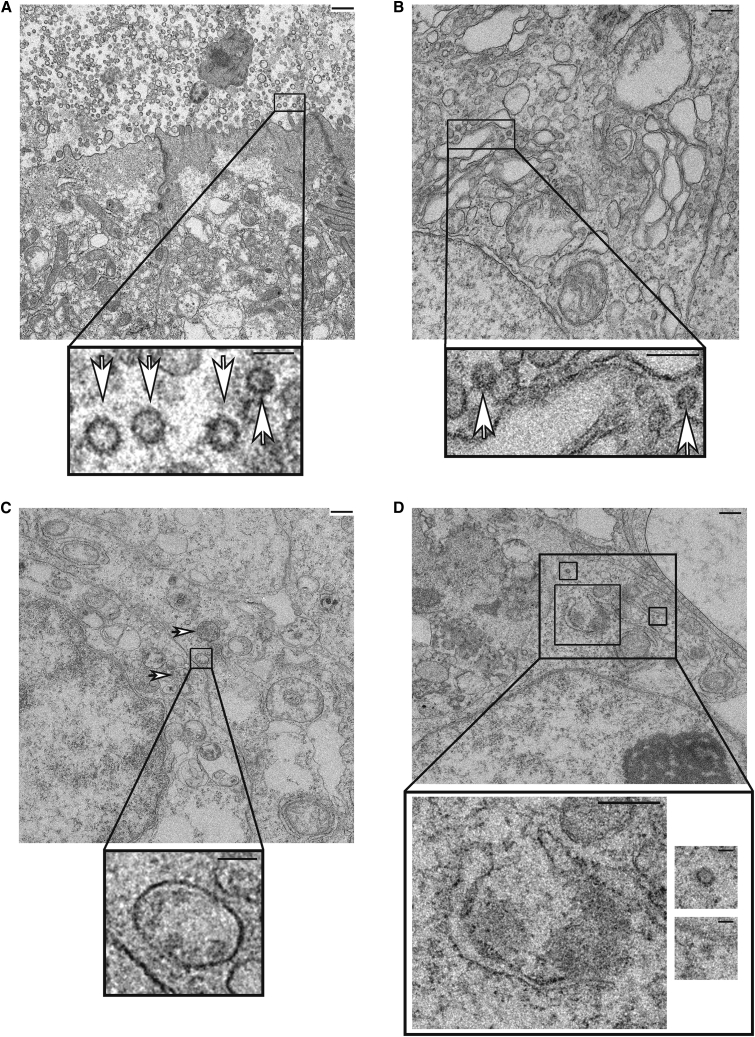
Transmission electron microscopy of both mock- and SARS-CoV-2-infected colonic and proximally patterned organoids was performed to examine the ultrastructural intracellular changes induced by infection. As expected, organoids contained cell types unique to the GI epithelium, including putative columnar microvilli-containing enterocytes (Figure S2C) and secretory cells (Figures S2D and S2E). Imaging of infected proximal organoids revealed the presence of characteristic coronaviral particles both within infected cells and adjacent to the enterocyte apical brush border (Figure 3A). Virions were also present within single bilayer and multilayered intracellular structures, at times budding off the Golgi, that contained convoluted membrane and putative viral replication complexes (Figures 3B and 3C) in both proximal and colonic organoids. As seen in analysis of COVID-19 patient specimens (Bradley et al., 2020), bilayered horseshoe-shaped structures were observed that may provide protected regions for viral replication (Figure 3D). No structures resembling virions or replication complexes were found in the mock-infected samples (Figure S2).
Figure 3.
Transmission Electron Microscopy of SARS-CoV-2-Infected Colonic and Proximal HIOS
Representative electron micrographs of (A) proximal HIO enterocytes infected with SARS-CoV-2 showing large numbers of coronavirus particles adjacent to the cell surface (inset, white arrowheads), (B) proximal HIO cytoplasm with budding virions (inset, white arrowheads), (C) infected colonic HIOs with intracellular replication complexes (white arrowheads) and viral particles within a single membrane vesicle (inset), and (D) cytoplasmic regions of viral replication with characteristic horseshoe structures and budding virions (inset). Scale bars, 500 nm (A, C, D large images), 200 nm (B, large image), or 125 nm (all insets); images representative of n = 3 replicate directed differentiation experiments. See also Figure S2.
SARS-CoV-2 Infection of HIOs Elicits an Inflammatory Response Common to Multiple Epithelial Cell Types
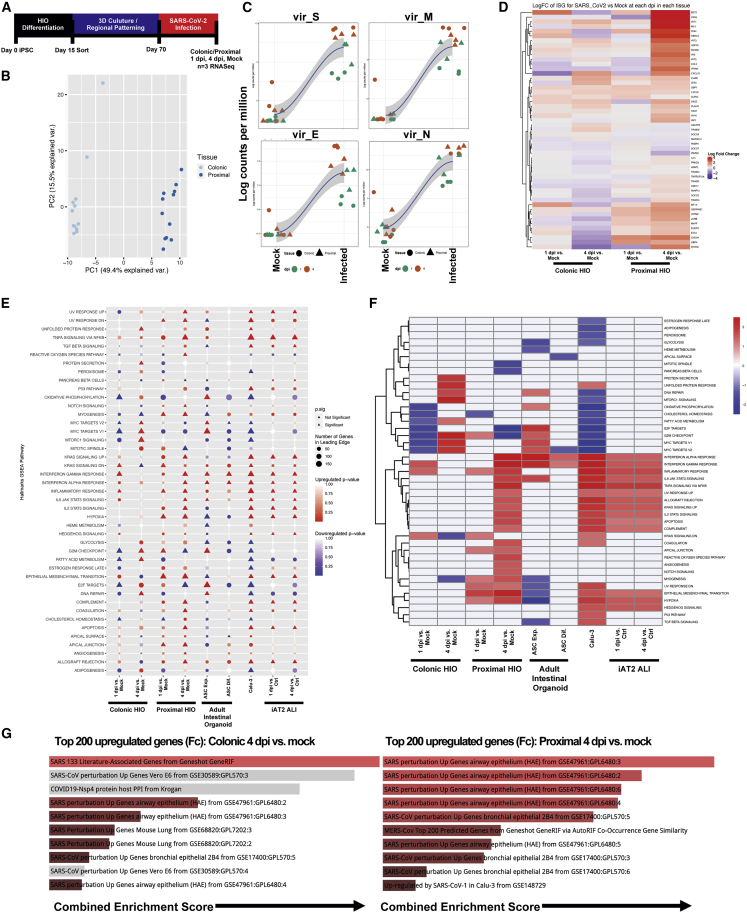
In order to define global transcriptional responses to viral infection, we harvested both mock- and SARS-CoV-2-infected proximal and colonic organoids at 1 and 4 dpi and performed bulk RNA-seq (n = 3 replicate directed differentiation experiments per condition tested at each time point) (Figure 4A). Principal component analysis (PCA) revealed that the primary differences in transcriptome across samples were between the tissue types (Figure 4B). However, all infected samples demonstrated profound upregulation of viral transcripts (Figures 4C and S3A–S3C), as well as significant global transcriptomic changes. Compared with corresponding mock-infected cells, there were 603 significantly differentially expressed genes (DEGs) in the infected proximal HIOs and 60 DEGs in the colonic HIOs at 1 dpi (false discovery rate [FDR] < 0.05). By 4 dpi, the number of DEGs increased considerably in both cell types, with 1,430 DEGs in the infected proximal HIOs compared with mock-infected cells and 4,339 DEGs in the colonic HIOs (FDR < 0.05). Among the changes, we observed a remarkable upregulation of a panel of interferon-stimulated genes that emerge 1 dpi but became particularly evident in the infected proximal HIOs at 4 dpi, highlighting a potentially differential response between proximal and colonic HIOs to SARS-CoV-2 infection (Figures 4D and 4E).
Figure 4.
Transcriptomic Response to SARS-CoV-2 Infection in Colonic and Proximal HIOs
(A) Experimental schematic of RNA-seq experiment, with n = 3 replicate directed differentiations per condition tested (colonic/proximal HIOs, 1 and 4 dpi with corresponding mock controls).
(B) PCA plot of mock and infected proximal and distal samples colored by tissue type, showing global transcriptomic variance between proximal and colonic HIOs.
(C) LOESS plots of key viral genes in mock (left) and infected (right) HIOs at 1 and 4 dpi.
(D) Unsupervised hierarchical clustered heatmap of differential gene expression of a subset of interferon-related genes as plotted by log fold change in HIOs at 1 and 4 dpi versus mock.
(E) GSEA (using hallmark gene sets) of top differentially expressed gene sets in colonic and proximal HIOs 1 and 4 dpi versus mock, compared with previously published SARS-CoV-2-related datasets, including ASC-derived intestinal organoids, Calu-3 cells, and iPSC-derived alveolar type 2 cells cultured at air-liquid interface. Statistical significance (p < 0.05) represented by triangles, size of shape represents the number of genes in the leading edge.
(F) Unsupervised hierarchical clustered heatmap of differentially expressed hallmark gene sets represented by fold change in HIOs 1 and 4 dpi versus mock and previously published data.
(G) GSEA of the top 200 upregulated genes at 4 dpi in colonic (left) and proximal (right) HIOs by log fold change versus mock using Enrichr, mapping to the top COVID-19-related gene sets. Length of bars indicates combined enrichment score. Red bars represent p < 0.05.
See also Figures S3 and S4.
Gene set enrichment analysis (GSEA) using hallmark gene sets was performed, revealing a significant upregulation of innate inflammatory pathways by 4 dpi (including IL2/IL6 JAK/STAT signaling genes) that again was more pronounced in proximal HIOs compared with colonic HIOs (Figure 4E). We next compared the transcriptomic response in our organoids with other published SARS-CoV-2-infected epithelial cells, including biopsy-obtained intestinal ASC-derived organoids (Lamers et al., 2020), Calu-3 cells (Blanco-Melo et al., 2020), and iPSC-derived lung epithelial cells (iAT2s) (Huang et al., 2020). Antiviral and inflammatory responses including the interferon gamma, interferon alpha, IL6-JAK-STAT3, and NF-κB pathways were observed across all epithelial cell types (Figure 4E). Our cells, particularly the proximal HIOs, recapitulated the inflammatory response to infection observed in iAT2 lung epithelial cells and ASC-derived intestinal organoids, with significant upregulation of interferon signaling in 4 dpi proximal HIOs (Figures 4E and 4F). In colonic HIOs, we observed a more pronounced upregulation of the unfolded protein response (UPR) and upregulation of genes downstream of NF-κB signaling, particularly compared with proximal HIOs, which displayed marked upregulation in GSEA pathways correlating to interferon alpha and gamma signaling (Figures 4E and 4F). In order to further examine the transcriptomic differences between both the baseline conditions as well as the response to infection and presence of increased cell death in proximal and colonic HIOs, we compared the top DEGs between the mock and infected conditions at both 1 and 4 dpi (Figures S4A and S4B). In mock-infected samples at 4 dpi, colonic HIOs expressed significantly higher levels (red dots) of markers specific to colonic epithelium, including SATB2, MUC2, and AQP8, while expressing significantly lower levels (blue dots) of markers specific to proximal small intestinal epithelium such as GATA4 and PDX1 (Figure S4C). Pan-intestinal markers, including CDX2, VIL1, and CDH17, were not differentially regulated between colonic and proximal HIOs, but were all highly expressed (Figure S4C). In SARS-CoV-2-infected colonic HIOs at 4 dpi, we observed upregulation of genes involved in the UPR (ATF6), the NF-κB signaling cascade (TRAF6, TNFRSF1A, MAP3K7), and general pro-inflammatory markers (HAVCR1, PLA2G2A) compared with proximal HIOs (Figure S4D). However, colonic HIOs downregulated canonical interferon-stimulated genes, including BST2, OASL, IFIT1, MX1, IFITM1, and IRF7, compared with proximal HIOs at 4 dpi (Figure S4D). Finally, we compared the top 200 upregulated genes by fold change (excluding viral transcripts) in colonic and proximal HIOs with other curated SARS-CoV-2-related publicly available datasets using Enrichr (Chen et al., 2013; Kuleshov et al., 2016). Strikingly, our HIOs recapitulated SARS-CoV-2-specific transcriptional responses observed in a variety of epithelial cell types, including large airway and bronchial epithelium (Figure 4F).
Discussion
The data presented here support the use of iPSC-derived HIOs as a robust and valuable model to study SARS-CoV-2 infection of various regional intestinal epithelia, and further support the notion of a conserved epithelial response to viral infection across cell and tissue type characterized by interferon signaling and downstream NF-κB activation. Notably, our ability to regionally pattern HIOs comprising entirely epithelial cells toward proximal or colonic-like lineages enables the study of differential host responses in a reductionist manner (Mithal et al., 2020). Our results demonstrate that both proximal and distal HIOs are readily permissive to productive SARS-CoV-2 infection, recapitulating prior work performed in other intestinal organoid systems (Lamers et al., 2020; Zang et al., 2020). SARS-CoV-2 infection did not induce any differences in intestinal-specific gene expression, or cellular phenotype. We also observed regional differences in the transcriptomic response to infection, with a more robust inflammatory signature in proximal HIOs as compared with colonic, which is of potential interest to clinicians caring for COVID-19 patients with pre-existing regionally specific GI diseases of increasing incidence and prevalence, such as Crohn, ulcerative colitis, or celiac (Nowak et al., 2020; Suárez-Fariñas et al., 2021). We observed upregulation of apoptosis-related genes in both the infected colonic and proximal HIOs compared with mock-infected cells at 4 dpi, including three specific markers for necroptosis (RIPK3, RIPK1, and MLKL) in the colonic HIOs, indicating the induction of cell death (Figure S4A). These results suggest that the cellular stress and inflammatory responses generated by SARS-CoV-2 infection in our HIOs could imply intestinal epithelial damage in vivo, a relevant finding pertaining to COVID-19 disease progression, in particular the diarrheal illness seen in these patients.
We observed significant upregulation of gene sets that correlated to the type 1 and type 2 interferon response pathways, which corroborates previous work implicating the importance of these pathways in SARS-CoV-2 host response both in vitro (Banerjee et al., 2020.; Huang et al., 2020; Lamers et al., 2020; Stanifer et al., 2020; Zhou et al., 2020a) and in vivo (Blanco-Melo et al., 2020; Hadjadj et al., 2020; Zhang et al., 2020). While our analysis did not show specific upregulation of a type III interferon annotated gene set as has been previously demonstrated (Stanifer et al., 2020; Vanderheiden et al., 2020), many of the genes in both the interferon alpha response and TNFA signaling via NF-κB gene sets (which were both significantly upregulated in infected HIOs) are also common to the type III interferon response. Notably, SARS-CoV-2 elicited a markedly delayed interferon response, in both colonic and proximal HIOs, with the bulk of interferon-related pathways only significantly upregulated at 4 dpi. This finding is of particular interest and could either be a result of a low infection rate at 1 dpi or direct viral suppression of the interferon response at early time points post infection. A delayed IFN response has also been reported for other in vitro infection models of SARS-CoV-2 (Blanco-Melo et al., 2020; Huang et al., 2020; Lei et al., 2020) as well as for SARS-CoV-1- and MERS-CoV-infected human airway epithelial cells and is a determinant of SARS disease severity (Channappanavar et al., 2016; Menachery et al., 2014). In COVID-19 patients, the impaired expression of innate immune mediators in SARS-CoV-2-infected epithelial cells across organs might contribute to the subsequent excessive inflammatory response, likely induced by migrating or tissue-resident immune cells (Hadjadj et al., 2020; Sanchez-Cerrillo et al., 2020). However, despite the observed pan-epithelial interferon and inflammatory response that has been confirmed by our study, there might be relevant organ-specific differences, leading to differential clinical presentations and outcomes in COVID-19 patients. We also observed significant upregulation of BST2 in response to infection, particularly in our proximal HIOs. Since BST2 was shown to have antiviral activity against both SARS-CoV-1 (Taylor et al., 2015) and SARS-CoV-2 (Stewart et al., 2021) by inhibiting viral release and, consequently, viral spread, this finding is of particular interest as it could explain regional-specific differences in response to infection within the GI tract. Our results warrant further investigation into the mechanisms of SARS-CoV-2-induced antagonism of innate immune responses both in general as well as within the GI tract, particularly in the context of COVID-19 patients harboring recently identified polymorphisms in interferon signaling that could affect their clinical course (Hadjadj et al., 2020; Livanos et al., 2020; Nobel et al., 2020). The GI tract is a site of major clinical relevance in SARS-CoV-2 infection, and our results demonstrate the potential implications of regional differences within the GI tract, as well as the significance of an epithelial-intrinsic inflammatory response to viral infection.
Our study is limited by the potential immaturity of our cells in the context of developmental milestones. Although our HIOs contain polarized, differentiated cell types specific to intestinal epithelium, many iPSC-derived cells and organoids resemble fetal-like cells, and may not fully recapitulate their adult counterparts. Further studies are clearly necessary in the context of the rapidly evolving situation surrounding the COVID-19 pandemic and would provide further insights into the proteomic and cell-signaling components of intestinal SARS-CoV-2 infection. These could also employ additional iPSC lines with different genetic backgrounds (as well as newly emerging strains of SARS-CoV-2) to investigate the role of specific genotype-phenotype interactions in the SARS-CoV-2 host response across various epithelial cell types. However, our iPSC-derived cells provide a readily scalable model for therapeutic screening purposes as well as a platform that is easily amenable to genetic manipulation in order to study genotype-specific disease susceptibility. The mesenchyme-free HIO system is also devoid of any mesenchymal or immune cells, removing a potential confounding variant and allowing for the reductionist capture of epithelial-intrinsic host response, and could lead to the development of protective therapies that ameliorate organ damage seen in COVID-19 patients. Furthermore, an iPSC-derived system could enable further studies involving additional epithelial and non-epithelial cell types (Yang et al., 2020), including iPSC-derived macrophages, endothelial cells, and other immune cells, enabling the study of host responses to SARS-CoV-2 and other infectious pathogens affecting the GI tract.
Experimental procedures
iPSC Lines and Maintenance
iPSC lines (BU1CG) were generated by our group (Mithal et al., 2020) from previously healthy individuals, and were shown to be karyotypically normal (46XY). All human subject studies were performed under signed consent and approved by the Boston University Institutional Review Board (IRB), protocol H-32506. BU1CG was maintained in feeder-free conditions using mTESR1 (StemCell Technologies) and passaged onto hESC Matrigel (Corning cat. no. 354277) coated, six-well tissue culture dishes (Corning) as per the manufacturer's instructions. For a detailed outline of standard iPSC culture and maintenance protocols, please visit http://www.bu.edu/dbin/stemcells/protocols.php.
Directed Differentiation of BU1CG to HIOs
Upon reaching >95% confluency, iPSCs were differentiated into HIOs using a protocol previously described by our group (Mithal et al., 2020). Briefly, confluent wells of iPSCs were dissociated into single-cell suspensions using Gentle Cell Dissociation Reagent (StemCell Technologies cat. no. 07174), and re-plated at a density of 2 × 106 cells per well of a Matrigel-coated six-well tissue culture plate in mTeSR1 supplemented with Y27632 (Tocris, 5 μM). Twenty-four hours post plating, cells were differentiated into definitive endoderm using the StemCell Technologies StemDiff Definitive Endoderm Kit (cat. no. 05110), as per manufacturer's instructions. At day 3, cells were split 1:3 as described above into new Matrigel-coated plates and incubated with DS/SB, containing Dorsomorphin (DS) (2 μM Stemgent, cat. no. 04-0024) and SB431542 (SB) (10 μM Tocris, cat. no. 1614) supplemented with Y27632 for 24 h, followed by DS/SB without Y27632 for 48 h. At day 6, cells were incubated in CB/RA containing CHIR99021 (C/CHIR) (3 μM, Tocris, cat. no. 4423), rhBMP4 (B) (10 ng/mL, R&D Systems, cat. no. 314-BP), and retinoic acid (RA) (100 nM, Sigma, cat. no. R2625-50MG). At day 14–15, cells were dissociated using 0.05% Trypsin-EDTA (ThermoFisher), and washed in Dulbecco's modified Eagle's medium (DMEM) with 20% FBS. Cells were then strained using a 40 -μm filter, spun at 300 × g for 5 min, and resuspended in FACS buffer containing 5 μM Y27632, and 10 nM Calcein Blue in DMSO (ThermoFisher, cat. no. C1429). Cells were then sorted to isolate the CDX2-eGFP+ populations using an operator-assisted MoFlo Astrios EQ (Beckman Coulter) at the Boston University Flow Cytometry Core Facility (FCCF). After sorting, cells were spun down and resuspended in 3D Matrigel (Corning 354234), in droplets of 50–100 μL, supplemented with either IM + CK (proximal, Intestinal Media + CHIR/KGF) or CK + DCI (colonic, CHIR/KGF + Dexamethasone, cAMP, and IBMX), at a density of 0.5–1 × 103 cells/μL, and plated on a pre-warmed 24-well tissue culture plate. After allowing the droplets to solidify for 20 min in a 37°C incubator, additional IM + CK or CK + DCI was added, supplemented with Y27632. After 3–4 days, fresh medium was added without Y27632, and with further medium replacement performed every 4–5 days, depending on confluency. Basal media for DS/SB, CB/RA, and CK + DCI consisted of complete serum-free differentiation medium (cSFDM), containing Iscove's Modified Dulbecco's Medium (IMDM; ThermoFisher) and Ham's F12 (ThermoFisher) with B27 supplement with retinoic acid (Invitrogen), N2 supplement (Invitrogen), 0.1% BSA fraction V (Invitrogen), monothioglycerol (Sigma), Glutamax (ThermoFisher), ascorbic acid (Sigma), and primocin. Basal media for IM + CK consisted of DMEM/F12 (ThermoFisher), B27 with retinoic acid (Invitrogen), N2 supplement (Invitrogen), Glutamax (ThermoFisher), and primocin. For media recipes, see Table S1, and for a comprehensive list of reagents and catalog numbers, including antibodies, see Table S2.
SARS-CoV-2 Propagation and Titration
SARS-CoV-2 stocks (isolate USA_WA1/2020, kindly provided by CDC's principal investigator Natalie Thornburg and the World Reference Center for Emerging Viruses and Arboviruses [WRCEVA]) were generated and purified as previously described (Huang et al., 2020). Briefly, SARS-CoV-2 was propagated in Vero E6 cells (ATCC CRL-1586) cultured in DMEM supplemented with 2% fetal calf serum (FCS), penicillin (50 U/mL), and streptomycin (50 mg/mL). Viral stocks were purified by ultracentrifugation through a 20% sucrose cushion at 80,000 × g for 2 h at 4°C (Huang et al., 2020). Viral titers were determined by tissue culture infectious dose 50 (TCID50) assay in Vero E6 cells using the Spearman Kärber algorithm. All work with SARS-CoV-2 was performed in the biosafety level 4 (BSL4) facility of the National Emerging Infectious Diseases Laboratories at Boston University following approved standard operating procedures. Pluripotent stem cell lines used in this study, along with maintenance standard operating procedures and directed differentiation protocols, are available from the CReM iPSC Repository at Boston University and Boston Medical Center and can be found at http://www.bu.edu/dbin/stemcells/.
SARS-CoV-2 Infection of HIOs
HIOs were infected with purified SARS-CoV-2 stock at the indicated MOI. One-hundred microliters of inoculum was prepared in CK + DCI or IM + CK media for colonic or proximal HIOs, respectively (or mock infected with medium only). To facilitate infection, HIOs were isolated from Matrigel. Briefly, medium was removed from wells containing HIOs, 0.5 mL of Cell Recovery Solution (Corning) was added to the cells in Matrigel, and plates were incubated at 4°C for 30 min. The dissolved Matrigel and cells in the Cell Recovery Solution were then transferred to 2-mL Sarstedt tubes. Cells were pelleted by low-speed centrifugation (300 × g, 5 min). Supernatant was removed and virus-free or virus-containing media were added to the corresponding tubes. Cell pellets were mechanically disrupted to facilitate viral access. HIOs were then incubated for 1 h at 37°C and 5% CO2. After the adsorption period, cells were pelleted by low-speed centrifugation (300 × g, 5 min) and the inoculum was removed. Cells were subsequently re-plated in Matrigel droplets with media and incubated at 37°C for the indicated times of infection.
At the time of harvest, medium was collected for viral titration as described above. Cell pellets were then recovered from Matrigel as described above. For immunofluorescence analysis, electron microscopy, and flow cytometry analysis, HIOs were fixed in 10% formalin for a minimal time of 6 h. For RNA-seq and qRT-PCR analysis, cells were lysed in TRIzol per manufacturer's specifications.
Transmission Electron Microscopy
Day 74 HIOs were infected with SARS-CoV-2 at an MOI of 0.4 or were mock infected. At 1 and 4 dpi, organoids were fixed and inactivated in 10% formalin for 6 h at 4°C and removed from the BSL-4 laboratory. The cells were washed with PBS and then post fixed in 1.5% osmium tetroxide (Polysciences) overnight at 4°C, and block stained in 1.5% uranyl acetate (Electron Microscopy Sciences [EMS]) for 1 h at room temperature (RT). The samples were dehydrated quickly through acetone on ice, from 70% to 80%–90%. The samples were then incubated two times in 100% acetone at RT for 10 min each, and in propylene oxide at RT for 15 min each. Finally, the samples were changed into EMbed 812 (EMS), left for 2 h at RT, changed into fresh EMbed 812, and left overnight at RT, after which they were embedded in fresh EMbed 812 and polymerized overnight at 60°C. Embedded samples were thin sectioned (70 nm) and grids were stained in 4% aqueous uranyl acetate for 10 min at RT followed by lead citrate for 10 min at RT. Transmission electron microscopy images were recorded on a Philips CM12 electron microscope operated at 100 kV using a TVIPS F216 CMOS camera with a pixel size of 0.85–3.80 nm.
Whole-Mount Immunostaining
Organoids were dissociated from their Matrigel droplets and fixed as described above. HIOs were then washed with PBS and blocked in 4% normal donkey serum (NDS) with 0.5% Triton X-100 (Sigma) for 30 min. After washing, HIOs were incubated in primary antibody diluted in 0.5% Triton X-100 and 4% NDS overnight at 4°C. Samples were washed in 4% NDS and incubated with secondary antibody from Jackson Immunoresearch (1:300 anti-rabbit IgG [H + L], 1:500 anti-chicken IgY, or anti-mouse IgG [H + L]) for 60 min at RT. HIOs were then washed again for 15 min with 0.5% Triton X-100 three times. Nuclei were stained with Hoechst dye (Thermo Fisher, 1:500). Whole organoids were then mounted with flouromont-G (Southern Biotech) on cavity slides and cover slipped. Stained HIOs were visualized with a Zeiss LSM 700 laser scanning confocal microscope.
Flow Cytometry
After infection and fixation, HIOs were manually dissociated into single-cell suspension, permeabilized with saponin buffer (Biolegend), and stained with SARS-CoV-2 nucleoprotein (N) antibody (Rockland, #200-401-A50, 1:1000) for 30 min at RT, followed by an incubation with donkey anti-rabbit IgG-AF647 (Thermo Fisher A-31573). Gating was performed with mock-infected stained cells. Samples were run on a Stratedigm S1000EXI instrument and analyzed with the FlowJo v10.6.2 software (FlowJo, Tree Star Inc). Shown FACS plots represent single cells based on forward-scatter/side-scatter gating.
RNA Isolation, cDNA Preparation, and qRT-PCR
HIOs were collected and lysed in TRIzol reagent (Thermo Fisher). RNA was extracted using the RNAeasy mini kit (QIAGEN) or following the manufacturer's protocol, respectively. cDNA was generated by reverse transcription using the SuperScript III First-Strand Synthesis System (Invitrogen cat. no. 18080093) as per the manufacturer's recommended parameters. RNA was quantified using a NanoDrop Lite Spectrophotometer (ThermoFisher) and input was standardized across all samples, to ensure normalized cDNA yields for downstream PCR applications. PCR was performed using TaqMan (Applied Biosystems) Master Mixes as per manufacturer's instructions, and the QuantStudio 6 Flex Real-Time 384 Well PCR System with barcoded 384-well plates. Relative fold change above undifferentiated iPSC or mock controls was determined by calculating the ΔΔCt, normalized to housekeeping gene GAPDH. For primer sequences, see the Table S2.
Library Preparation and Bulk mRNA Sequencing of SARS-CoV-2-Infected HIOs
RNA was extracted and prepared as described above from n = 3 replicate directed differentiations per condition tested (day 93–97 Colonic/Proximal HIOs, 1 and 4 dpi with corresponding mock controls). mRNA was isolated from each sample using magnetic bead-based poly(A) selection (New England BioLabs #6420), followed by synthesis of cDNA fragments. The products were end paired and PCR amplified to create each final cDNA library. Sequencing of pooled libraries was done using a NextSeq 500 (Illumina). The quality of the raw data was assessed using FastQC v.0.11.7. The sequence reads were aligned to a combination of the human genome reference (GRCh38) and the SARS CoV2 virus reference (NC_045512) with a tdTomato marker using STAR v.2.6.0c (Dobin et al., 2013). Counts per gene were summarized using the featureCounts function from the subread package v.1.6.2.
The matrix of counts per gene per sample was then analyzed using the limma/voom normalization (limma v. 3.42.2, edgeR v.3.25.10) (Ritchie et al., 2015). After exploratory data analysis (Glimma v. 1.11.1) (Su et al., 2017), contrasts for differential expression testing were done for each SARS-CoV2-infected sample versus mock (controls) at each time point (dpi) and in each tissue (colonic or proximal) separately. Differential expression testing was also conducted for each of the two tissues to compare the gene expression between time points and to investigate the time-specific effects in response to infection in each tissue separately. Functional predictions were performed using fgsea v.1.12.0 for gene set analysis (Korotkevich et al., 2019). The full dataset from this RNA-seq experiment can be found on Gene Expression Omnibus (GEO), accession number GEO: GSE159201.
Statistical Analysis
Statistical methods relevant to each figure are outlined in the figure legends. Briefly, unpaired, two-tailed Student's t tests were used to compare quantitative analyses comprising two groups of n = 3 or more independent biological replicates (separate differentiations). Further specifics about the replicates used in each experiment are available in the figure legends. p value annotations on graphs are as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. A p value threshold of 0.05 was used to determine significance. Data were reported with either standard error of the mean or standard deviation (SD) as specified in the figure legends.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts, Gustavo Mostoslavsky and Elke Mühlberger (gmostosl@bu.edu, muehlber@bu.edu).
Data and Code Availability
The RNA-seq data are available via the GEO, accession number GEO:GSE159201. This study did not generate new unique reagents. All previously published reagents are available with a completed materials transfer agreement. Pluripotent stem cell lines used in this study, along with maintenance standard operating procedures and directed differentiation protocols, are available from the CReM iPSC Repository at Boston University and Boston Medical Center and can be found at http://www.bu.edu/dbin/stemcells/.
Author contributions
A.M. performed directed differentiations and post-infection analysis, including qRT-PCR, flow cytometry, and immuno-staining, and wrote the first draft of the manuscript. A.J.H. performed all experiments at BSL-4, including viral infections, harvesting of infected organoids and RNA extraction, and viral stock maintenance and viral titering, and also edited the manuscript. J.V.L. and C.V.M. performed bioinformatics analyses. J.O. assisted with BSL-4 experiments. A.H. and E.B. prepared samples for and performed transmission electron microscopy. E.M. and G.M. designed the study and edited the manuscript.
Acknowledgments
We would like to thank Brian Tilton of the Boston University Medical Campus (BUMC) Flow Cytometry Core; Yuriy Alekseyev and Ashley LeClerc of the BUMC Microarray and Sequencing Resource Core; Vickery Trinkaus-Randall for assistance with confocal microscopy; Greg Miller, Andrew McCracken, Mitchell White, and Baylee Heiden for their invaluable technical support maintaining smooth laboratory operations throughout the pandemic; and George Murphy for his generous donation of the SARS-CoV-2 N qRT-PCR probes. This work was supported by Evergrande MassCPR, Fast Grants, and NIH NCATS grant UL1TR001430 awards to E.M. A.M. is supported by the Kilachand Multicellular Design Program at Boston University. G.M. is supported by NIH grants N0175N92020C00005 and 1R01DA051889-01.
Published: April 13, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.02.019.
Contributor Information
Elke Mühlberger, Email: muehlber@bu.edu.
Gustavo Mostoslavsky, Email: gmostosl@bu.edu.
Supplemental information
Information
References
- Abo K.M., Ma L., Matte T., Huang J., Alysandratos K.D., Werder R.B., Mithal A., Beermann M.L., Lindstrom-Vautrin J., Mostoslavsky G. Human iPSC-derived alveolar and airway epithelial cells can be cultured at air-liquid interface and express SARS-CoV-2 host factors. bioRxiv. 2020 2020.2006.2003.132639. [Google Scholar]
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmunzer B.J., Spitzer R.L., Foster L.D., Merchant A.A., Howard E.F., Patel V.A., West M.K., Qayed E., Nustas R., Zakaria A. Digestive manifestations in patients hospitalized with COVID-19. Clin. Gastroenterol. Hepatol. 2020;31371-9:1542–3565. doi: 10.1016/j.cgh.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2020;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., Sigl V., Hanada T., Hanada R., Lipinski S. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekman R.M., Hume A.J., Goel R.K., Abo K.M., Huang J., Blum B.C., Werder R.B., Suder E.L., Paul I., Phanse S. Actionable cytopathogenic host responses of human alveolar type 2 cells to SARS-CoV-2. Mol. Cell. 2021;81:212. doi: 10.1016/j.molcel.2020.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell. 2020;27:962–973.e7. doi: 10.1016/j.stem.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019 060012. [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livanos A.E., Jha D., Cossarini F., Gonzalez-Reiche A.S., Tokuyama M., Aydillo T., Parigi T.L., Ramos I., Dunleavy K., Lee B. Gastrointestinal involvement attenuates COVID-19 severity and mortality. medRxiv. 2020 [Google Scholar]
- Mahlakoiv T., Ritz D., Mordstein M., DeDiego M.L., Enjuanes L., Muller M.A., Drosten C., Staeheli P. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J. Gen. Virol. 2012;93:2601–2605. doi: 10.1099/vir.0.046284-0. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Eisfeld A.J., Schafer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5 doi: 10.1128/mBio.01174-14. e01174-01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithal A., Capilla A., Heinze D., Berical A., Villacorta-Martin C., Vedaie M., Jacob A., Abo K., Szymaniak A., Peasley M. Generation of mesenchyme free intestinal organoids from human induced pluripotent stem cells. Nat. Commun. 2020;11:215. doi: 10.1038/s41467-019-13916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel Y.R., Phipps M., Zucker J., Lebwohl B., Wang T.C., Sobieszczyk M.E., Freedberg D.E. Gastrointestinal symptoms and coronavirus disease 2019: a case-control study from the United States. Gastroenterology. 2020;159:373–375.e2. doi: 10.1053/j.gastro.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak J.K., Lindstrom J.C., Kalla R., Ricanek P., Halfvarson J., Satsangi J. Age, inflammation, and disease location are critical determinants of intestinal expression of SARS-CoV-2 receptor ACE2 and TMPRSS2 in inflammatory bowel disease. Gastroenterology. 2020;159:1151–1154.e2. doi: 10.1053/j.gastro.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cerrillo I., Landete P., Aldave B., Sanchez-Alonso S., Sanchez-Azofra A., Marcos-Jimenez A., Avalos E., Alcaraz-Serna A., de Los Santos I., Mateu-Albero T. COVID-19 severity associates with pulmonary redistribution of CD1c+ DC and inflammatory transitional and nonclassical monocytes. J. Clin. Invest. 2020;130:6290–6300. doi: 10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanifer M.L., Kee C., Cortese M., Zumaran C.M., Triana S., Mukenhirn M., Kraeusslich H.G., Alexandrov T., Bartenschlager R., Boulant S. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart H., Johansen K.H., McGovern N., Palmulli R., Carnell G.W., Heeney J.L., Okkenhaug K., Firth A.E., Peden A.A., Edgar J.R. SARS-CoV-2 spike downregulates tetherin to enhance viral spread. bioRxiv. 2021 [Google Scholar]
- Su S., Law C.W., Ah-Cann C., Asselin-Labat M.L., Blewitt M.E., Ritchie M.E. Glimma: interactive graphics for gene expression analysis. Bioinformatics. 2017;33:2050–2052. doi: 10.1093/bioinformatics/btx094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M., Tokuyama M., Wei G., Huang R., Livanos A., Jha D., Levescot A., Irizar H., Kosoy R., Cording S. Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-CoV-2 related disease. Gastroenterology. 2021;160:287–301.e20. doi: 10.1053/j.gastro.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.K., Coleman C.M., Postel S., Sisk J.M., Bernbaum J.G., Venkataraman T., Sundberg E.J., Frieman M.B. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J. Virol. 2015;89:11820–11833. doi: 10.1128/JVI.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.F., Lo A.W. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2) J. Pathol. 2004;203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala M.H., Porvari K.S., Kellokumpu S., Kyllonen A.P., Vihko P.T. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J. Pathol. 2001;193:134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94:e00985-20. doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh Y.K., Zuo T., Lui G.C., Zhang F., Liu Q., Li A.Y., Chung A.C., Cheung C.P., Tso E.Y., Fung K.S. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Zhan H., Zhang F., Liu Q., Tso E.Y.K., Lui G.C.Y., Chen N., Li A., Lu W., Chan F.K.L. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159:1302–1310.e5. doi: 10.1053/j.gastro.2020.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information
Data Availability Statement
The RNA-seq data are available via the GEO, accession number GEO:GSE159201. This study did not generate new unique reagents. All previously published reagents are available with a completed materials transfer agreement. Pluripotent stem cell lines used in this study, along with maintenance standard operating procedures and directed differentiation protocols, are available from the CReM iPSC Repository at Boston University and Boston Medical Center and can be found at http://www.bu.edu/dbin/stemcells/.