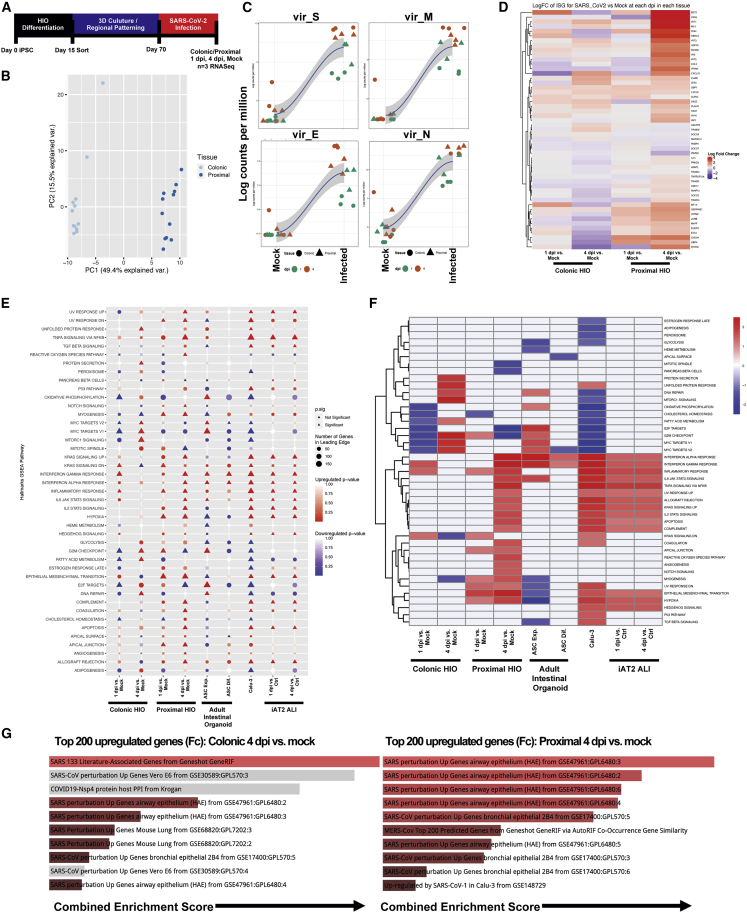
Figure 4.
Transcriptomic Response to SARS-CoV-2 Infection in Colonic and Proximal HIOs
(A) Experimental schematic of RNA-seq experiment, with n = 3 replicate directed differentiations per condition tested (colonic/proximal HIOs, 1 and 4 dpi with corresponding mock controls).
(B) PCA plot of mock and infected proximal and distal samples colored by tissue type, showing global transcriptomic variance between proximal and colonic HIOs.
(C) LOESS plots of key viral genes in mock (left) and infected (right) HIOs at 1 and 4 dpi.
(D) Unsupervised hierarchical clustered heatmap of differential gene expression of a subset of interferon-related genes as plotted by log fold change in HIOs at 1 and 4 dpi versus mock.
(E) GSEA (using hallmark gene sets) of top differentially expressed gene sets in colonic and proximal HIOs 1 and 4 dpi versus mock, compared with previously published SARS-CoV-2-related datasets, including ASC-derived intestinal organoids, Calu-3 cells, and iPSC-derived alveolar type 2 cells cultured at air-liquid interface. Statistical significance (p < 0.05) represented by triangles, size of shape represents the number of genes in the leading edge.
(F) Unsupervised hierarchical clustered heatmap of differentially expressed hallmark gene sets represented by fold change in HIOs 1 and 4 dpi versus mock and previously published data.
(G) GSEA of the top 200 upregulated genes at 4 dpi in colonic (left) and proximal (right) HIOs by log fold change versus mock using Enrichr, mapping to the top COVID-19-related gene sets. Length of bars indicates combined enrichment score. Red bars represent p < 0.05.
See also Figures S3 and S4.