Abstract
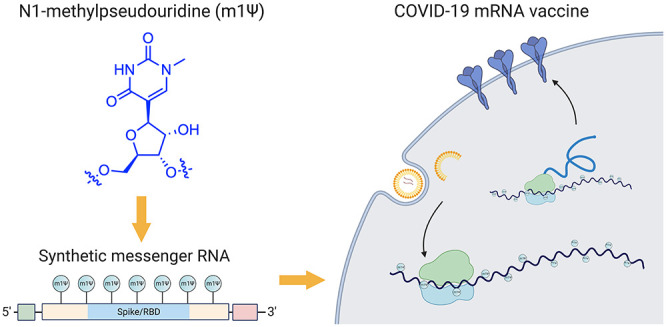
The novel coronavirus SARS-CoV-2, the cause of the COVID-19 pandemic, has inspired one of the most efficient vaccine development campaigns in human history. A key aspect of COVID-19 mRNA vaccines is the use of the modified nucleobase N1-methylpseudouridine (m1Ψ) to increase their effectiveness. In this Outlook, we summarize the development and function of m1Ψ in synthetic mRNAs. By demystifying how a novel element within these medicines works, we aim to foster understanding and highlight future opportunities for chemical innovation.
Short abstract
This article summarizes the development and function of N1-methylpseudouridine, an RNA modification used to increase the safety and efficacy of messenger RNA vaccines targeting COVID-19.
Introduction
On December 11, 2020, the U.S. Food and Drug Administration (FDA) issued the first emergency use authorization (EUA) for a vaccine to prevent COVID-19, a disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 Approval of a second COVID-19 vaccine followed 1 week later.2 These approvals represent a public health breakthrough, providing the first protective measures against the largest global pandemic to strike in over 100 years, and were the first fruit of a vaccine development process akin in scope and urgency to the famed Manhattan Project. These two vaccines are also notable for being the first FDA-approved therapeutics to use a novel therapeutic platform: synthetic mRNA (mRNA).
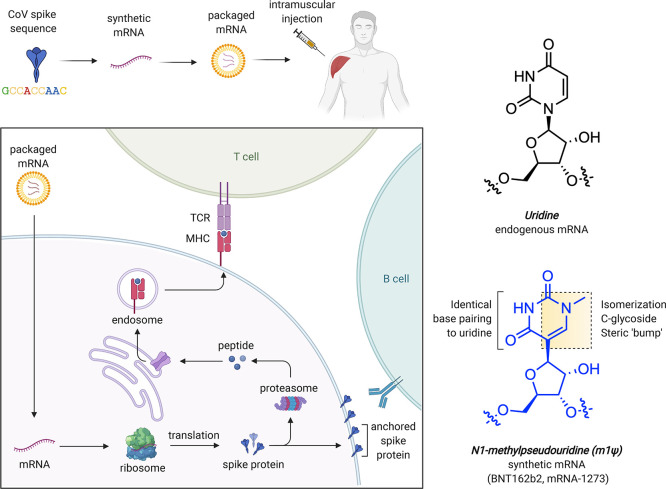
Messenger RNAs are used in every cell of our body, where they serve the central relay between the instructions of the genome and protein production. Synthetic mRNAs tap into this same natural process but are designed to encode proteins with therapeutic effects.3 The COVID-19 mRNA vaccines produce a full-length SARS-CoV-2 spike protein with two mutations (K986P and V987P) that ensure it remains in an antigenically favorable prefusion conformation.4,5 Upon injection, mRNA is taken up by muscle and infiltrating immune cells that use it to produce spike protein (Figure 1a). A transmembrane anchor causes the spike protein to be displayed on the cell surface, allowing it to be recognized by the immune system. This triggers the production of antibodies and T-cells that protect against natural infection and prevent serious disease. Since synthetic mRNAs produce only a single component of the SARS-CoV-2 genome, they cannot cause COVID-19. It is also important to note these vaccines are nonreplicating mRNAs that naturally decompose and do not integrate into genomes. Detailed descriptions of the development and characterization of these vaccines can be found in the primary literature reporting them as well as several excellent reviews.3,6−8
Figure 1.
(a) mRNA-based COVID-19 vaccine strategy. (b) Structural features of uridine and m1Ψ. TCR = T-cell receptor. MHC = major histocompatibility complex.
The chemical components of mRNA vaccines are pleasantly unremarkable, consisting primarily of RNA plus “water, salt, sugar, and fat,” with two notable exceptions. The first is the lipid nanoparticles that encapsulate the mRNA and facilitate its delivery, which are excellently reviewed elsewhere.9 The second is the non-natural RNA nucleobase N1-methylpseudouridine (m1Ψ; Figure 1b), which enhances immune evasion and protein production. In this Outlook, we briefly review the development and function of m1Ψ in synthetic mRNA. By demystifying how a critical component of these new medicines work, we hope to help foster their acceptance and highlight future areas for chemical innovation.
Primary Structure of the COVID-19 mRNA Vaccines
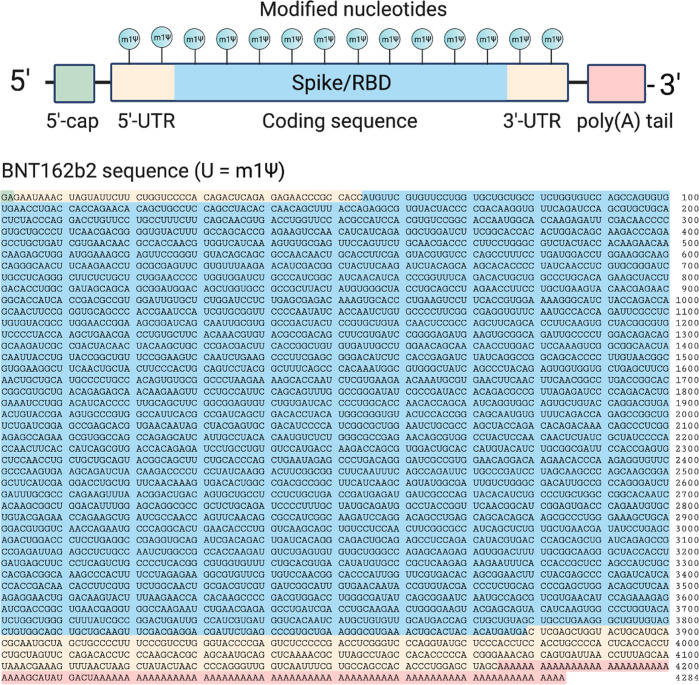
The two approved COVID-19 mRNA vaccines are marketed by Pfizer-BioNTech (BNT162b2; trade name: Comirnaty; generic name: tozinameran) and Moderna (mRNA-1273). The sequence of the former has been disclosed (Figure 2).10 The active payload of the Pfizer-BioNTech vaccine is a 4284 nucleotide linear sequence of RNA consisting of five main elements:11
A 5′-cap (m7(3′OMeG)(5′)ppp(5′)(2′OMeA)pG, commonly referred to as trinucleotide “cap 1”) that helps recruit the ribosome and protect the RNA from degradation.12
A 5′-untranslated region (UTR) derived from the human α-globin mRNA with an optimized Kozak sequence that helps drive high levels of translation from the correct start codon.13
A codon-optimized coding sequence that specifies production of the transmembrane-anchored immunogenic SARS-CoV-2 spike glycoprotein.
A 3′-UTR conisting of two sequences derived from the amino-terminal enhancer of split mRNA and the mitochondrial encoded 12S rRNA, which aids high levels of protein expression by stabilizing the RNA.14
An unusual 3′-terminus consisting of two segmented poly(adenosine) tracts. The poly(adenosine) stretches increase mRNA stability, while the segmented structure helps reduce unwanted recombination during plasmid production.15
Figure 2.
Top: Design elements found in synthetic mRNA therapeutics. Bottom: Sequence of the COVID-19 mRNA vaccine tozinameran (BNT162b2) from Pfizer/BioNTech. Green: 5′-cap. Yellow: 5′- and 3′-UTR sequences. Blue: SARS-CoV-2 spike glycoprotein coding sequence. Red: Segmented poly(A) tail.
The swift design of these vaccines has been deservedly celebrated.16,17 However, it is important to gently push back on the narrative that this process was hurried, which may invite skepticism. Each of the elements above were highly intentional choices that in many cases reflect decades of fundamental research in the RNA biology field.18,19 Below, we first review how these modified mRNAs are made, followed by an analysis of the modification’s biological effects.
Incorporation of N1-Methylpseudouridine into mRNA Vaccines
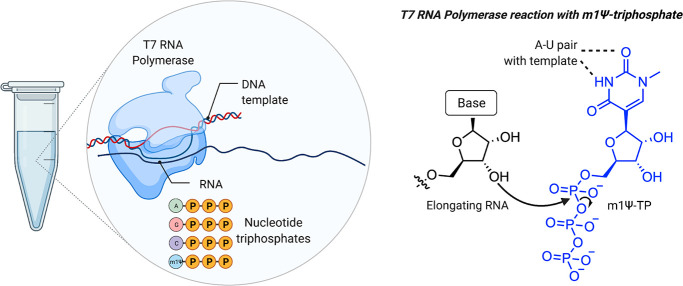
To evaluate the design above requires first overcoming a technical challenge: how does one produce (at scale) a synthetic mRNA with a linear sequence far longer than can be chemically synthesized while simultaneously preserving the flexibility to incorporate modified nucleobases such as m1Ψ? The answer has been to take a cue from nature and make them enzymatically (Figure 3). This approach takes advantage of the fact that DNA (which is far easier to synthesize than RNA) can be stitched together into large synthetic fragments. These fragments are used to construct plasmids, in which the code for the COVID-19 vaccine is placed downstream of a sequence that promotes its transcription into mRNA by recombinant T7 RNA polymerase. By incubating these plasmids with T7 polymerase and nucleotide triphosphates (NTPs), high yields of mRNA are produced. Decades of research have characterized T7 polymerase as a remarkable enzyme, which can produce RNAs longer than 20 000 nucleotides without making an error.20,21 Another feature of T7 polymerase is its tolerance for non-natural NTPs. Over 50 years ago, Goldberg and Rabinowitz demonstrated that RNA polymerases can incorporate pseudouridine triphosphate into RNA.22 In one early study (which makes one quite thankful for the Sigma-Aldrich catalogue), pseudouridine was isolated from 20 L of urine donated by patients with leukemia, polycythemia, or gout, converted to a radiolabeled triphosphate by a mixed chemoenzymatic approach, and found to replace uridine in RNA during in vitro transcription when UTP was omitted.23 Early studies of T7 RNA polymerase found it was also permissive of modified NTPs that do not alter base pairing,24 and this strategy has since been applied to many different bases.25−29 One caveat to this enzymatic approach is that it replaces the natural nucleobase with a non-natural residue homogeneously; in the case of BNT162b2, every uridine residue in the mRNA is replaced with m1Ψ. This means to be useful in a therapeutic mRNA, a modified nucleobase must be compatible with all of its functional elements, including UTRs and the coding sequence recognized by the ribosome. With this understanding of the primary sequence of modified mRNA vaccines and how they are produced, we can proceed to a discussion of what they do.
Figure 3.
Production of m1Ψ mRNAs by in vitro transcription. Left: Components of in vitro transcription reaction. Right: Incorporation of m1Ψ-triphosphate into RNA is guided by m1Ψ’s ability to form a canonical base pair with adenine of the DNA template in the T7 RNA polymerase active site.
N1-Methylpseudouridine Reduces mRNA Immunogenicity
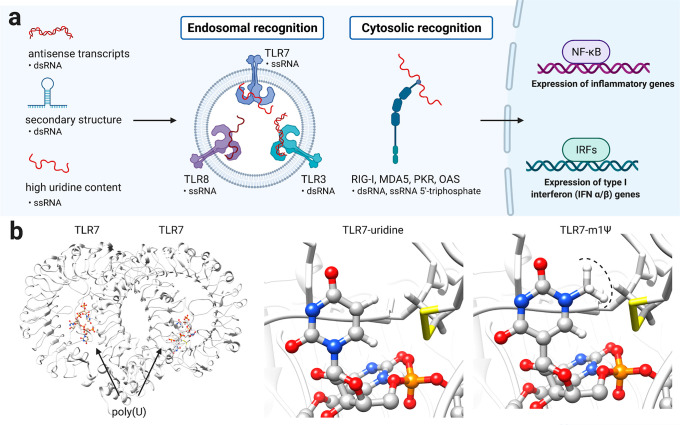
Early studies showed that synthetic mRNAs entrapped in cationic lipid vesicles can be transfected into cultured cells.30 When injected into mouse muscle, reporter mRNAs produced detectable proteins for weeks.31 However, a challenge to application of these agents as vaccines and protein replacement therapies was their immunogenicity. Cells contain a variety of pattern recognition receptors whose natural role is to identify and respond to viral RNAs by inducing downstream signaling. These include the endosomal receptors TLR3, TLR7, and TLR8, which recognize double- and single-stranded RNA, and the cytosolic receptors RIG-I and MDA-5, which recognize double-stranded and 5′-triphosphate-modified RNA. While induction of an immune response is theoretically a positive attribute for a vaccine, uncontrolled immune activation can lead to allergic reactions and anaphylactic shock. Furthermore, at a molecular level, overstimulation of immune signaling is known to silence protein translation, with the potential outcome of limiting antigen expression and vaccine efficacy. A breakthrough came from the fundamental studies of Kariko and co-workers, who showed that many modifications naturally found in human RNA such as pseudouridine, thiouridine, and 5-methylcytidine reduce its immunostimulatory potential.32 This inspired follow-up studies demonstrating that these same nucleobase modifications could increase protein production from synthetic mRNAs33−35 and be applied in many applications, including the generation of induced pluripotent stem cells.36−38 Further development of this concept led to m1Ψ, which in mRNA was found to increase protein output while decreasing TLR3 activation.39 The ability of m1Ψ and related modifications to reduce the immunogenicity of synthetic mRNA has been attributed to at least three mechanisms (Figure 4):
Figure 4.
(a) Activation of innate immune response by mRNA secondary structures (b) Structure of the single-stranded RNA sensor TLR7 in complex with a polyuridine (poly(U)) ligand (PDB ID: 5GMF). Replacing uridine with m1Ψ demonstrates the steric incompatibility of the modified nucleobase with TLR7 binding and immune activation.
Reduced synthesis of antisense RNA: Under high-yielding conditions, T7 RNA polymerase sometimes uses the RNA it has produced to “self-prime”, leading to the synthesis of small amounts of duplexed antisense mRNA (Figure 4).40 Removal of these double-stranded RNA impurities by chromatography does not eliminate differences in immunogenicity observed between m1Ψ-modified and unmodified RNAs but does reduce it.41 Other studies have also found that using base-modified NTPs yields noninflammatory mRNAs without the need for purification.34,42 This suggests that using the non-natural NTP for RNA synthesis may disfavor this side product.
Altering interaction with RNA secondary structure: In addition to antisense impurities, mRNA can form secondary structures such as hairpins that may be recognized by immune receptors such as TLR3 and RIG-I (Figure 4).43 Incorporation of modified bases has the potential to reduce these recognition events by altering secondary structure and protein/double-stranded RNA interactions. In the related C-glycoside pseudouridine, isomerization shifts the structural equilibrium of the nucleotide toward a C3′-endo ribose sugar and an anti orientation of the base, a conformation that favors helicity and stacking.44−46 Consistent with this, a recent study used chemical probing reagents to find evidence that RNAs containing m1Ψ and uridine form distinct secondary structures.47 Modified nucleotides have also been found to reduce the ability of mRNAs to propagate immune signaling through RIG-I, indicative of their ability to influence protein–RNA interactions.48
Altering interaction with single-stranded RNA immune receptors: In immune cells, single-stranded poly(uridine) RNA is one of the most potent inducers of interferon and is sensed by TLR7.49,50 To define whether m1Ψ alters immune recognition of single-stranded RNA, a recent study assessed the ability of RNAs containing this species to activate inflammatory gene expression.41,47 To ensure any differences were not due to double-stranded RNA, the authors employed a mouse model where the immune response to these structures was silenced. Even in the absence of double-stranded RNA sensing, m1Ψ RNAs were less inflammatory than those containing canonical uridine. This suggests the altered hydrogen bonding face and steric “bump” presented by m1Ψ disrupts the interaction of immune sensors such as TLR7 with single-stranded segments of synthetic mRNA (Figure 4c).
It is important to note that in many studies, the specific contributions of each of these mechanisms to mRNA immunogenicity have not been explicitly defined. In such cases, an mRNA modification may be exerting its activity by altering antisense transcript synthesis, mRNA structure, immune recognition, or some combination thereof.
Vaccines often require coadministration of adjuvants, which are agents that prime the immune system to respond to an antigen of interest. In the case of tozinameran and mRNA-1273, this role appears to be fulfilled by the lipid nanoparticle, which can be tailored to predictably activate the immune response via mechanisms that do not halt protein production.51−53 Separating the adjuvant from the nucleic acid component of the vaccine reduces the chance that the mRNA sequence composition may influence vaccine efficacy. This potentially increases the strategy’s generality and also opens the door to other applications, such as treatment of autoimmune disorders54 and therapeutic protein replacement.55
Interestingly, at least two groups have reported that pseudouridine, the natural analogue of m1Ψ, does not measurably alter mRNA immunogenicity in vivo(56) and that many of the benefits of m1Ψ can be obtained by simply engineering a synthetic mRNA’s sequence to limit the use of uridine-containing codons.57 A comparative analysis of codons used in tozinameran relative to the spike glycoprotein encoded by the SARS-CoV-2 genome observes a disproportionate depletion of uridine residues, indicative of sequence engineering (Figure S1). In the context of the COVID-19 vaccine, the relative effects of sequence engineering and m1Ψ incorporation on the immunogenic mechanisms specified above remains to be reported.
N1-Methylpseudouridine Can Alter mRNA Translation
The ultimate purpose of an mRNA medicine is to express a therapeutic protein. Thus, m1Ψ and other modified bases have been explored for their ability to facilitate the translation of mRNA into protein via the ribosome. These studies are naturally intertwined with those above, as immune activation can limit translation by shutting down the ribosome and activating ribonucleases that degrade mRNA. Consistent with this, in the initial report where m1Ψ-containing mRNA was found to drive high levels of protein production, this was attributed in part to its ability to blunt TLR3 activation.39 To decouple translation and immune activation, Svitkin and co-workers analyzed the translation of m1Ψ mRNAs in a cell-free translation system.58 They observed that incorporation of m1Ψ increases the size and abundance of polysomes, leading them to propose that the more rapid translation initiation and slower elongation of m1Ψ mRNAs may coordinately increase their half-life as well as induce productive interactions with the ribosome. These studies provided the first evidence that m1Ψ may directly impact mRNA translation.
Natural RNA modifications are known to be context-dependent.59 This means they can exert different effects on different RNAs. Those effects may also be dependent on where in the RNA they lie (e.g., UTR, coding sequence).60,61 Two studies have examined the context-dependence of m1Ψ in a high-throughput fashion (Figure 5).47,62 In the first, Sample et al. used RNA sequencing of polysomes to compare how a library of uridine and m1Ψ mRNAs containing 280 000 different 5′-UTRs was loaded onto the ribosome. Across all sequences tested, ribosome loading was found to be anticorrelated with predicted mean free energy. This is consistent with the classical view that structured 5′-UTRs can repress translation.63 However, this anticorrelation was stronger for m1Ψ than uridine, indicating that by stabilizing RNA structure, the modified base may actually decrease protein production in these contexts.46,64 A second study by Mauger et al. examined the relationship between m1Ψ, RNA structure, and protein production in even greater detail. They evaluated modified (m1Ψ, pseudouridine, methoxyuridine) and unmodified (uridine) mRNAs across multiple synonymous versions of three different reporters, amounting to over 150 synthetic mRNAs in total. Within this library, modified and unmodified mRNAs were found to exhibit distinct “fingerprints” of codon optimality. Assuming uridine and m1Ψ are decoded similarly by the ribosome, this suggested that a feature other than codon optimality is responsible for tuning synthetic mRNA translation. To examine the potential role of structure in this process, the authors used a biochemical probing technique (SHAPE-MaP)65 to study modified and unmodified mRNAs. As in the case of 5′-UTRs, it was found that m1Ψ stabilizes structure. Further studies provided support for a model in which secondary structure in the coding sequence, which can be enforced by m1Ψ, may increase mRNA functional half-life independent of codon optimality.47,62
Figure 5.
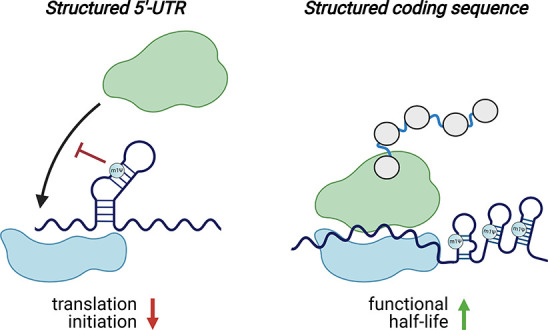
m1Ψ exerts context-dependent effects on translation. Left: m1Ψ-dependent enforcement of secondary structure in the 5′-UTR of synthetic mRNAs can inhibit translation initiation. Right: m1Ψ-dependent enforcement of secondary structure in the coding sequences of synthetic mRNAs can increase their functional half-life. Note: While m1Ψ is homogeneously incorporated throughout synthetic mRNA vaccines, in these illustrations, m1Ψ is only specified in duplexes to emphasize its potential to influence mRNA structure.
One important aspect revealed by these studies is that m1Ψ is not a panacea for protein production. While for most mRNA sequences m1Ψ performed as well or better than uridine, in some it performed worse. Similar observations have been made for pseudouridine, which in one study was found to be incompatible with protein output from mRNAs containing structured viral internal ribosomal entry sites in their 5′-UTR region.57 The efficient translation of many different m1Ψ-containing mRNAs suggests that the secondary structures induced by this modification do not activate immune sensors. This may reflect their small size or greater dynamics relative to the stable duplexes found in classic TLR3 agonists such as poly(I:C) or the intrinsic ability of m1Ψ to impede the protein–mRNA interactions responsible for immune activation.
Conclusions
The shock of the COVID-19 pandemic mobilized the biomedical research community on an unprecedented scale and enabled the most rapid vaccine production process in human history. This success also presents a unique challenge to scientific communication, which is how to highlight the decades of fundamental research that underlie these medicines. In this Outlook, we describe for a scientific lay audience the development and application of m1Ψ, a chemical component of COVID-19 mRNA vaccines. The modified nucleobase helps cloak mRNA vaccines from the immune system, limiting their undesired immune stimulation, and in certain circumstances may also enhance the synthesis of antigens by the protein-producing machinery of the cell. This allows these vaccines to tap into the natural process of mRNA translation without triggering harmful side effects such as anaphylaxis.
In light of the current concern over emerging SARS-CoV-2 variants, it is worth highlighting how synthetic mRNAs are being developed for use in personalized cancer immunotherapy.66,67 In this approach, clinicians remove a tumor, sequence it to identify coding mutations, and use this information to design custom mRNAs that express those mutant peptides at high levels, which helps train the immune system to selectively attack tumor tissue.68 In other words, synthetic mRNA platforms have been built with the express purpose of rapidly addressing newly discovered mutations. This bodes well for the potential of these medicines to be reconfigured to combat emerging viral strains and suggests one unexpected legacy of this pandemic may be to accelerate the use of synthetic mRNAs in cancer treatment.
Finally, our review of m1Ψ highlights future areas where chemical innovation may help extend the reach of therapeutic mRNAs. First, while the modular nature of mRNA vaccines has led to considerable enthusiasm, the combinatorial space of elements that contribute to their activity (including caps, coding sequence, codons, UTRs, and modifications) is massive in scale, and relatively few RNA modifications have been comparatively evaluated in a systematic manner. High-throughput approaches will be critical to help define this space and develop optimized agents.69 The exploration of novel nucleobases may be also be aided by efficient routes to nucleoside triphosphates70 as well as biological insights arising from the recent renaissance in the study of endogenous mRNA modifications.71,72 The production of novel mRNA therapies may also be aided by the evolution of RNA polymerases with improved synthetic properties such as expanded nucleobase tolerance or a reduced production of antisense transcripts. The successful engineering of DNA polymerases for genome sequencing speaks to the feasibility and potential impact of this goal.
Almost 60 years ago in “Meditations in an Emergency” the poet Frank O’Hara wrote, “I am needed by things as the sky must be above the earth./And lately, so great has their anxiety become, I can spare myself little sleep.” O’Hara’s passage resonates with our current era and the tremendous strain felt by patients, families, and healthcare providers during this pandemic. The nucleobase m1Ψ, a “modification in an emergency”, provides an example of how contemplation can also lead to intervention, offering hope and rest in a time of crisis.
Acknowledgments
The authors thank members of the Meier lab as well as Igor Ulitsky (Weizmann Institute of Science) and Lu Chen (National Center for Advancing Translation Sciences, NIH) for helpful discussions. Portions of graphics were adapted from “COVID-19 DNA-Based Vaccine” and “TLR Signaling Pathway” by BioRender.com (2020). This work was supported by the National Institutes of Health, National Cancer Institute, Center for Cancer Research (ZIA BC011488).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00197.
Full sequences for SARS-CoV-2 spike glycoprotein, tozinameran, and Supplementary Figures (PDF)
The authors declare no competing financial interest.
A putative sequence for the Moderna vaccine was reported while this manuscript was in production.73 Although this sequence has not been validated, the initial report does include the statement that, “the RNAs that are now a part of the human ecosystem and that are likely to appear in numerous other high throughput RNA-seq studies in which a fraction of the individuals may have previously been vaccinated.” In our view this is likely erroneous, as there is no evidence for long-term detection of mRNA vaccines in vaccinated individuals by RNA-seq. Indeed, all experimental evidence to date supports the view that synthetic mRNAs are efficiently destroyed by the body in the days following after vaccination.
Supplementary Material
References
- Pfizer-BioNTech COVID-19 Vaccine EUA Letter of Authorization; U.S. Food and Drug Administration, 2021.
- Moderna COVID-19 Vaccine EUA Letter of Authorization; U.S. Food and Drug Administration, 2021.
- Sahin U.; Karikó K.; Türeci Ö. mRNA-Based Therapeutics — Developing a New Class of Drugs. Nat. Rev. Drug Discovery 2014, 13 (13), 759–780. 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Yang C.; Xu X.-F.; Xu W.; Liu S.-W. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41 (9), 1141–1149. 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L.; Goldsmith J. A.; Schaub J. M.; DiVenere A. M.; Kuo H.-C.; Javanmardi K.; Le K. C.; Wrapp D.; Lee A. G.; Liu Y.; et al. Structure-Based Design of Prefusion-Stabilized SARS-CoV-2 Spikes. Science 2020, 369 (6510), 1501–1505. 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares-Fernández S.; Lacroix C.; Exposito J.-Y.; Verrier B. Tailoring mRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol. Med. 2020, 26 (3), 311–323. 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Pardi N.; Hogan M. J.; Weissman D. Recent Advances in mRNA Vaccine Technology. Curr. Opin. Immunol. 2020, 65, 14–20. 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Chung Y. H.; Beiss V.; Fiering S. N.; Steinmetz N. F. COVID-19 Vaccine Frontrunners and Their Nanotechnology Design. ACS Nano 2020, 14 (10), 12522–12537. 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- Kulkarni J. A.; Darjuan M. M.; Mercer J. E.; Chen S.; van der Meel R.; Thewalt J. L.; Tam Y. Y. C.; Cullis P. R. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 2018, 12 (5), 4787–4795. 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- World Health Organization: International Nonproprietary Names Programme . Messenger RNA Encoding the Full-Length SARS-CoV-2 Spike Glycoprotein; 11889; 2020.
- Vogel A. B.; Kanevsky I.; Che Y.; Swanson K. A.; Muik A.; Vormehr M.; Kranz L. M.; Walzer K. C.; Hein S.; Güler A.; et al. BNT162b Vaccines Protect Rhesus Macaques from SARS-CoV-2. Nature 2021. 10.1038/s41586-021-03275-y [DOI] [PubMed] [Google Scholar]
- Henderson J. M.; Ujita A.; Hill E.; Yousif-Rosales S.; Smith C.; Ko N.; McReynolds T.; Cabral C. R.; Escamilla-Powers J. R.; Houston M. E. Cap 1 Messenger RNA Synthesis with Co-Transcriptional CleanCap® Analog by In Vitro Transcription. Curr. Protoc 2021, 1 (2), e39 10.1002/cpz1.39. [DOI] [PubMed] [Google Scholar]
- Babendure J. R.; Babendure J. L.; Ding J.-H.; Tsien R. Y. Control of Mammalian Translation by mRNA Structure near Caps. RNA 2006, 12 (5), 851–861. 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandini von Niessen A. G.; Poleganov M. A.; Rechner C.; Plaschke A.; Kranz L. M.; Fesser S.; Diken M.; Löwer M.; Vallazza B.; Beissert T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019, 27 (4), 824–836. 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepotec Z.; Geiger J.; Plank C.; Aneja M. K.; Rudolph C. Segmented poly(A) Tails Significantly Reduce Recombination of Plasmid DNA without Affecting mRNA Translation Efficiency or Half-Life. RNA 2019, 25 (4), 507–518. 10.1261/rna.069286.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi J. COVID-19 and mRNA Vaccines-First Large Test for a New Approach. JAMA 2020, 324 (12), 1125–1127. 10.1001/jama.2020.16866. [DOI] [PubMed] [Google Scholar]
- Collins F.Celebrating the Gift of COVID-19 Vaccines. https://directorsblog.nih.gov/2020/12/22/celebrating-the-gift-of-covid-19-vaccines/.
- Su S.; Du L.; Jiang S. Learning from the Past: Development of Safe and Effective COVID-19 Vaccines. Nat. Rev. Microbiol. 2021, 19, 211–219. 10.1038/s41579-020-00462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B. S. Rapid COVID-19 Vaccine Development. Science 2020, 368 (6494), 945–946. 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Chamberlin M.; McGrath J.; Waskell L. New RNA Polymerase from Escherichia Coli Infected with Bacteriophage T7. Nature 1970, 228 (5268), 227–231. 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M.; Kingston R.; Gilman M.; Wiggs J.; deVera A. Isolation of Bacterial and Bacteriophage RNA Polymerases and Their Use in Synthesis of RNA in Vitro. Methods Enzymol. 1983, 101, 540–568. 10.1016/0076-6879(83)01037-X. [DOI] [PubMed] [Google Scholar]
- Goldberg I. H.; Rabinowitz M. The Incorporation of 5-Ribosyluracil Triphosphate into RNA in Nuclear Extracts of Mammalian Cells. Biochem. Biophys. Res. Commun. 1961, 6, 394–398. 10.1016/0006-291X(61)90152-8. [DOI] [PubMed] [Google Scholar]
- Goldberg I. H.; Rabinowitz M. Comparative Utilization of Pseudouridine Triphosphate and Uridine Triphosphate by Ribonucleic Acid Polymerase. J. Biol. Chem. 1963, 238, 1793–1800. 10.1016/S0021-9258(18)81139-5. [DOI] [PubMed] [Google Scholar]
- Milligan J. F.; Uhlenbeck O. C. Synthesis of Small RNAs Using T7 RNA Polymerase. Methods Enzymol. 1989, 180, 51–62. 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Li B.; Luo X.; Dong Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjugate Chem. 2016, 27 (3), 849–853. 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- Sinclair W. R.; Arango D.; Shrimp J. H.; Zengeya T. T.; Thomas J. M.; Montgomery D. C.; Fox S. D.; Andresson T.; Oberdoerffer S.; Meier J. L. Profiling Cytidine Acetylation with Specific Affinity and Reactivity. ACS Chem. Biol. 2017, 12 (12), 2922–2926. 10.1021/acschembio.7b00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto M.; Meyer A. J.; Hirao I.; Ellington A. D. Genetic Alphabet Expansion Transcription Generating Functional RNA Molecules Containing a Five-Letter Alphabet Including Modified Unnatural and Natural Base Nucleotides by Thermostable T7 RNA Polymerase Variants. Chem. Commun. 2017, 53 (91), 12309–12312. 10.1039/C7CC06661A. [DOI] [PubMed] [Google Scholar]
- Milisavljevič N.; Perlíková P.; Pohl R.; Hocek M. Enzymatic Synthesis of Base-Modified RNA by T7 RNA Polymerase. A Systematic Study and Comparison of 5-Substituted Pyrimidine and 7-Substituted 7-Deazapurine Nucleoside Triphosphates as Substrates. Org. Biomol. Chem. 2018, 16 (32), 5800–5807. 10.1039/C8OB01498A. [DOI] [PubMed] [Google Scholar]
- Hopkins P. A.; McCoy L. S.; Tor Y. Enzymatic Incorporation and Utilization of an Emissive 6-Azauridine. Org. Biomol. Chem. 2017, 15 (3), 684–690. 10.1039/C6OB02080A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. W.; Felgner P. L.; Verma I. M. Cationic Liposome-Mediated RNA Transfection. Proc. Natl. Acad. Sci. U. S. A. 1989, 86 (16), 6077–6081. 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J. A.; Malone R. W.; Williams P.; Chong W.; Acsadi G.; Jani A.; Felgner P. L. Direct Gene Transfer into Mouse Muscle in Vivo. Science 1990, 247 (4949), 1465–1468. 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Karikó K.; Buckstein M.; Ni H.; Weissman D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23 (2), 165–175. 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karikó K.; Muramatsu H.; Welsh F. A.; Ludwig J.; Kato H.; Akira S.; Weissman D. Incorporation of Pseudouridine into mRNA Yields Superior Nonimmunogenic Vector with Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16 (11), 1833–1840. 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K.; Muramatsu H.; Ludwig J.; Weissman D. Generating the Optimal mRNA for Therapy: HPLC Purification Eliminates Immune Activation and Improves Translation of Nucleoside-Modified, Protein-Encoding mRNA. Nucleic Acids Res. 2011, 39 (21), e142 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson B. R.; Muramatsu H.; Nallagatla S. R.; Bevilacqua P. C.; Sansing L. H.; Weissman D.; Karikó K. Incorporation of Pseudouridine into mRNA Enhances Translation by Diminishing PKR Activation. Nucleic Acids Res. 2010, 38 (17), 5884–5892. 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L.; Manos P. D.; Ahfeldt T.; Loh Y.-H.; Li H.; Lau F.; Ebina W.; Mandal P. K.; Smith Z. D.; Meissner A.; Daley G. Q.; Brack A. S.; Collins J. J.; Cowan C.; Schlaeger T. M.; Rossi D. J. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell 2010, 7 (5), 618–630. 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewska L.; Kitada T.; Endo K.; Siciliano V.; Stillo B.; Saito H.; Weiss R. Mammalian Synthetic Circuits with RNA Binding Proteins for RNA-Only Delivery. Nat. Biotechnol. 2015, 33 (8), 839–841. 10.1038/nbt.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormann M. S. D.; Hasenpusch G.; Aneja M. K.; Nica G.; Flemmer A. W.; Herber-Jonat S.; Huppmann M.; Mays L. E.; Illenyi M.; Schams A.; et al. Expression of Therapeutic Proteins after Delivery of Chemically Modified mRNA in Mice. Nat. Biotechnol. 2011, 29 (2), 154–157. 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- Andries O.; Mc Cafferty S.; De Smedt S. C.; Weiss R.; Sanders N. N.; Kitada T. N(1)-Methylpseudouridine-Incorporated mRNA Outperforms Pseudouridine-Incorporated mRNA by Providing Enhanced Protein Expression and Reduced Immunogenicity in Mammalian Cell Lines and Mice. J. Controlled Release 2015, 217, 337–344. 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Mu X.; Greenwald E.; Ahmad S.; Hur S. An Origin of the Immunogenicity of in Vitro Transcribed RNA. Nucleic Acids Res. 2018, 46 (10), 5239–5249. 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.; Sorensen E. W.; Mintri S.; Rabideau A. E.; Zheng W.; Besin G.; Khatwani N.; Su S. V.; Miracco E. J.; Issa W. J.; Hoge S.; Stanton M. G.; Joyal J. L.; et al. Impact of mRNA Chemistry and Manufacturing Process on Innate Immune Activation. Sci. Adv. 2020, 6 (26), eaaz6893 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan S.; Azizian K. T.; Haque A. K. M. A.; Henderson J. M.; Hendel A.; Shore S.; Antony J. S.; Hogrefe R. I.; Kormann M. S. D.; Porteus M. H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther.--Nucleic Acids 2018, 12, 530–542. 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K.; Ni H.; Capodici J.; Lamphier M.; Weissman D. mRNA Is an Endogenous Ligand for Toll-like Receptor 3. J. Biol. Chem. 2004, 279 (13), 12542–12550. 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- Davis D. R. Stabilization of RNA Stacking by Pseudouridine. Nucleic Acids Res. 1995, 23 (24), 5020–5026. 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-C.; Herath J.; Wang T. H.-H.; Chow C. S. Synthesis and Solution Conformation Studies of 3-Substituted Uridine and Pseudouridine Derivatives. Bioorg. Med. Chem. 2008, 16 (5), 2676–2686. 10.1016/j.bmc.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E.; Malgowska M.; Lisowiec J.; Turner D. H.; Gdaniec Z.; Kierzek R. The Contribution of Pseudouridine to Stabilities and Structure of RNAs. Nucleic Acids Res. 2014, 42 (5), 3492–3501. 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger D. M.; Cabral B. J.; Presnyak V.; Su S. V.; Reid D. W.; Goodman B.; Link K.; Khatwani N.; Reynders J.; Moore M. J.; et al. mRNA Structure Regulates Protein Expression through Changes in Functional Half-Life. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (48), 24075–24083. 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin A. F.; Wang C.; Marcotrigiano J.; Gehrke L. RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 2016, 7 (5), e00833-16 10.1128/mBio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F.; Hemmi H.; Hochrein H.; Ampenberger F.; Kirschning C.; Akira S.; Lipford G.; Wagner H.; Bauer S. Species-Specific Recognition of Single-Stranded RNA via Toll-like Receptor 7 and 8. Science 2004, 303 (5663), 1526–1529. 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Diebold S. S.; Massacrier C.; Akira S.; Paturel C.; Morel Y.; Reis e Sousa C. Nucleic Acid Agonists for Toll-like Receptor 7 Are Defined by the Presence of Uridine Ribonucleotides. Eur. J. Immunol. 2006, 36 (12), 3256–3267. 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- Alving C. R.; Beck Z.; Matyas G. R.; Rao M. Liposomal Adjuvants for Human Vaccines. Expert Opin. Drug Delivery 2016, 13 (6), 807–816. 10.1517/17425247.2016.1151871. [DOI] [PubMed] [Google Scholar]
- Perrie Y.; Crofts F.; Devitt A.; Griffiths H. R.; Kastner E.; Nadella V. Designing Liposomal Adjuvants for the next Generation of Vaccines. Adv. Drug Delivery Rev. 2016, 99, 85–96. 10.1016/j.addr.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Calcagnile S.; Zuccotti G. V. The Virosomal Adjuvanted Influenza Vaccine. Expert Opin. Biol. Ther. 2010, 10 (2), 191–200. 10.1517/14712590903431014. [DOI] [PubMed] [Google Scholar]
- Krienke C.; Kolb L.; Diken E.; Streuber M.; Kirchhoff S.; Bukur T.; Akilli-Öztürk Ö.; Kranz L. M.; Berger H.; Petschenka J.; et al. A Noninflammatory mRNA Vaccine for Treatment of Experimental Autoimmune Encephalomyelitis. Science 2021, 371 (6525), 145–153. 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- An D.; Schneller J. L.; Frassetto A.; Liang S.; Zhu X.; Park J.-S.; Theisen M.; Hong S.-J.; Zhou J.; Rajendran R.; et al. Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia. Cell Rep. 2018, 24 (9), 2520. 10.1016/j.celrep.2018.08.049. [DOI] [PubMed] [Google Scholar]
- Kauffman K. J.; Mir F. F.; Jhunjhunwala S.; Kaczmarek J. C.; Hurtado J. E.; Yang J. H.; Webber M. J.; Kowalski P. S.; Heartlein M. W.; DeRosa F.; et al. Efficacy and Immunogenicity of Unmodified and Pseudouridine-Modified mRNA Delivered Systemically with Lipid Nanoparticles in Vivo. Biomaterials 2016, 109, 78–87. 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thess A.; Grund S.; Mui B. L.; Hope M. J.; Baumhof P.; Fotin-Mleczek M.; Schlake T. Sequence-Engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015, 23 (9), 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y. V.; Cheng Y. M.; Chakraborty T.; Presnyak V.; John M.; Sonenberg N. N1-Methyl-Pseudouridine in mRNA Enhances Translation through eIF2α-Dependent and Independent Mechanisms by Increasing Ribosome Density. Nucleic Acids Res. 2017, 45 (10), 6023–6036. 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boo S. H.; Kim Y. K. The Emerging Role of RNA Modifications in the Regulation of mRNA Stability. Exp. Mol. Med. 2020, 52 (3), 400–408. 10.1038/s12276-020-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H.; Wei J.; He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74 (4), 640–650. 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I. A.; Evans M. E.; Pan T.; He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169 (7), 1187–1200. 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample P. J.; Wang B.; Reid D. W.; Presnyak V.; McFadyen I. J.; Morris D. R.; Seelig G. Human 5′ UTR Design and Variant Effect Prediction from a Massively Parallel Translation Assay. Nat. Biotechnol. 2019, 37 (7), 803–809. 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Regulation of Translation via mRNA Structure in Prokaryotes and Eukaryotes. Gene 2005, 361, 13–37. 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Mustoe A. M.; Corley M.; Laederach A.; Weeks K. M. Messenger RNA Structure Regulates Translation Initiation: A Mechanism Exploited from Bacteria to Humans. Biochemistry 2018, 57 (26), 3537–3539. 10.1021/acs.biochem.8b00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried N. A.; Busan S.; Rice G. M.; Nelson J. A. E.; Weeks K. M. RNA Motif Discovery by SHAPE and Mutational Profiling (SHAPE-MaP). Nat. Methods 2014, 11 (9), 959–965. 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara M. A.; Nair S. K.; Holl E. K. RNA-Based Vaccines in Cancer Immunotherapy. J. Immunol. Res. 2015, 2015, 794528. 10.1155/2015/794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara M. L.; Persano F.; Persano S. Advances in Lipid Nanoparticles for mRNA-Based Cancer Immunotherapy. Front. Chem. 2020, 8, 589959. 10.3389/fchem.2020.589959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor F.; Berraondo P.; Etxeberria I.; Frederick J.; Sahin U.; Gilboa E.; Melero I. An RNA Toolbox for Cancer Immunotherapy. Nat. Rev. Drug Discovery 2018, 17 (10), 751–767. 10.1038/nrd.2018.132. [DOI] [PubMed] [Google Scholar]
- Asrani K. H.; Farelli J. D.; Stahley M. R.; Miller R. L.; Cheng C. J.; Subramanian R. R.; Brown J. M. Optimization of mRNA Untranslated Regions for Improved Expression of Therapeutic mRNA. RNA Biol. 2018, 15 (6), 756–762. 10.1080/15476286.2018.1450054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J.-Y.; Bala S.; Ngor A. K.; Yik E. J.; Chaput J. C. P(V) Reagents for the Scalable Synthesis of Natural and Modified Nucleoside Triphosphates. J. Am. Chem. Soc. 2019, 141 (34), 13286–13289. 10.1021/jacs.9b04728. [DOI] [PubMed] [Google Scholar]
- Eyler D. E.; Franco M. K.; Batool Z.; Wu M. Z.; Dubuke M. L.; Dobosz-Bartoszek M.; Jones J. D.; Polikanov Y. S.; Roy B.; Koutmou K. S. Pseudouridinylation of mRNA Coding Sequences Alters Translation. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (46), 23068–23074. 10.1073/pnas.1821754116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T. M.; Martinez N. M.; Schaening C.; Su A.; Bell T. A.; Zinshteyn B.; Gilbert W. V. mRNA Structure Determines Modification by Pseudouridine Synthase 1. Nat. Chem. Biol. 2019, 15 (10), 966–974. 10.1038/s41589-019-0353-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.; McCoy M.; Artiles K.; Ilbay O.; Fire A.; Nadeau K.; Park H.; Betts B.; Boyd S.; Hoh R.; Shoura M.. Assemblies-of-putative-SARS-CoV2-spike-encoding-mRNA-sequences-for-vaccines-BNT-162b2-and-mRNA-1273. https://github.com/NAalytics/Assemblies-of-putative-SARS-CoV2-spike-encoding-mRNA-sequences-for-vaccines-BNT-162b2-and-mRNA-1273/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.