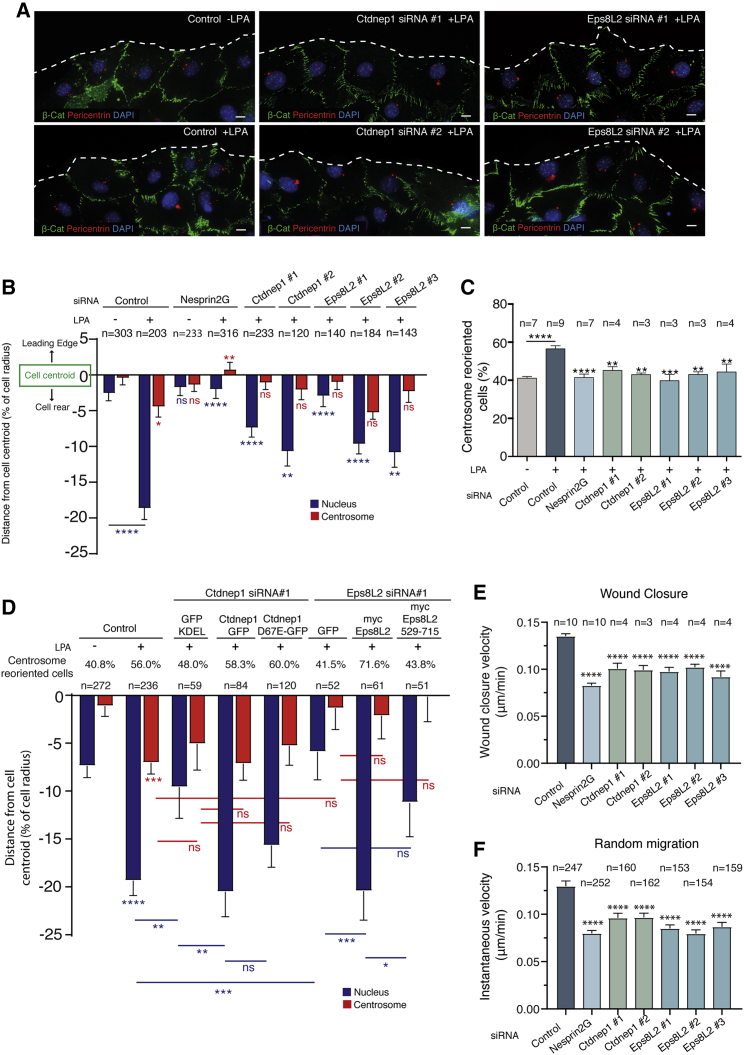
Figure 1.
Ctdnep1 and Eps8L2 are required for nuclear positioning and cell migration in fibroblasts
(A) Representative images of wound-edge 3T3 fibroblasts with or without LPA stimulation in control, Ctdnep1, and Eps8L2 siRNAs stained for β-catenin (green, cell contacts), pericentrin (red, centrosome), and DAPI (blue, nucleus). The dashed white lines mark the wound edge.
(B) Average positions of the nucleus (blue) and centrosome (red) relative to the cell centroid in cells treated with control, Nesprin2G, Ctdnep1, and Eps8L2 siRNAs.
(C) Percentage of oriented cells in the conditions analyzed in (B).
(D) Average positions of the nucleus (blue) and centrosome (red) in cells treated with Ctdnep1 or Eps8L2 siRNAs and microinjected with KDEL-GFP, Ctdnep1-GFP, Ctdnep1_D67E-GFP (no phosphatase activity), GFP, myc-Eps8L2, or myc-Eps8L2_529-715.
(E and F) Average wound closure velocity during wound closure migration (E) and instantaneous cell velocity in random migration (F) in cells treated with control, Nesprin2G, Ctdnep1, and Eps8L2 siRNAs.
Data are represented as mean ± SEM. The n value means number of analyzed cells (B, D, and F) or number of experiments (C and E) with >50 cells analyzed per experiment. Statistics was performed by unpaired t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Scale bars 10μm. See also Figure S1.