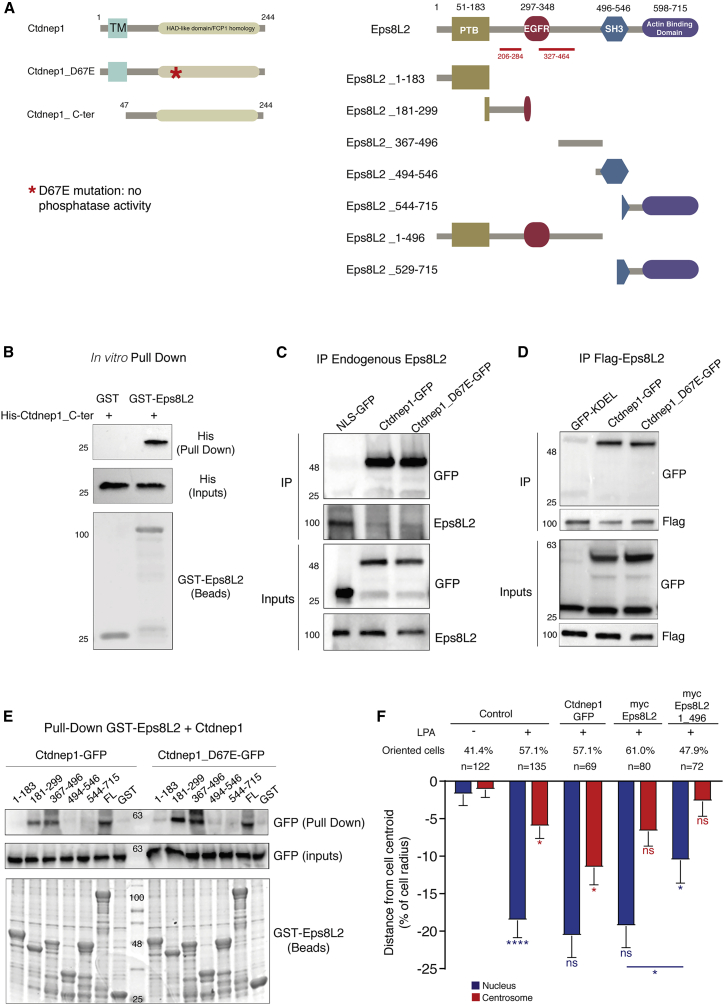
Figure 2.
Ctdnep1 and Eps8L2 directly interact
(A) Schematic representation of Ctdnep1, Ctdnep1_D67E (red asterisk denotes D67E mutation), Ctdnep1_C-ter (Ctdnep1 without the transmembrane domain), and Eps8L2 proteins showing their different protein domains and fragments used in this work. TM indicates transmembrane domain. Red lines in Eps8L2 indicate the regions of interaction with Ctdnep1 obtained in the Y2H assay.
(B) In vitro pull-down of recombinant GST-Eps8L2 bound to glutathione agarose beads with purified recombinant His-Ctdnep1_C-ter.
(C) Co-immunoprecipitation of endogenous Eps8L2 from SRKB cells with NLS-GFP, Ctdnep1-GFP, and Ctdnpe1_D67E-GFP from transfected U2OS cells.
(D) Co-immunoprecipitation of FLAG-Eps8L2 overexpressed in U2OS cells and posteriorly incubated with GFP-KDEL, Ctdnep1-GFP, and Ctdnep1_D67E-GFP overexpressed in U2OS cells.
(E) Pull-down assay of recombinant GST-Eps8L2 and its different fragments bound to glutathione agarose beads with Ctdnep1-GFP and Ctdnep1_D67E-GFP overexpressed in U2OS cells.
(F) Average positions of the nucleus (blue) and centrosome (red) in wild-type cells microinjected with Ctdnep1-GFP, myc-Eps8L2, or myc-Eps8L2_1-496. The n value means number of analyzed cells.
Data are represented as mean ± SEM. Statistics was performed by unpaired t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S2.