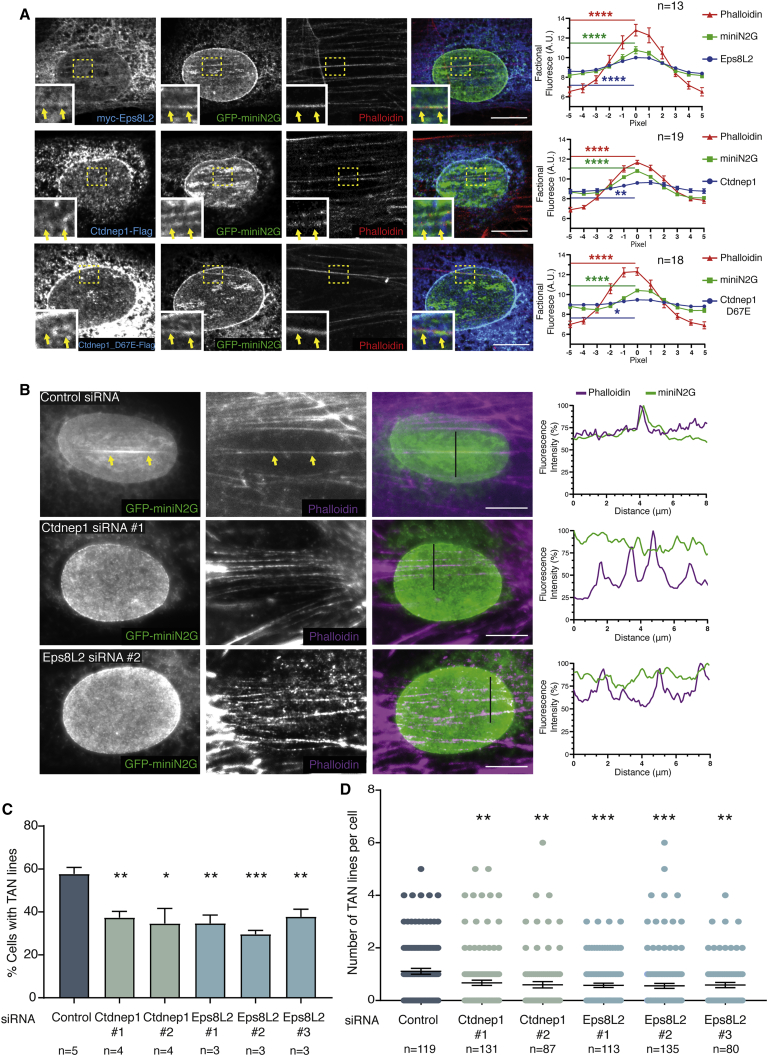
Figure 3.
Ctdnep1 and Eps8L2 are required for TAN lines formation
(A) Wound-edge fibroblast stimulated with LPA and microinjected with GFP-miniN2G and myc-Eps8L2 (top panels), Ctdnep1-FLAG (middle panels), and Ctdnep1_D67E-FLAG (bottom panels). Cells were stained for GFP (green), phalloidin (red), myc (blue), and FLAG (blue). Dashed yellow squares indicate the regions of the insets shown in the bottom left corner. Colocalization of miniN2G and actin (phalloidin) in linear arrays at the nuclear envelope is indicated by yellow arrows in the insets. Quantification of the colocalization between miniNesprin2G, phalloidin, and Eps8L2 or Ctdnep1 is shown in the right plots. For the quantification, an 11-pixels-size box (width) was designed with the TAN line centered in the central pixel in order to measure the intensity for each channel and for each row of pixels (see STAR methods). The n value means number of TAN lines analyzed.
(B) Wound-edge fibroblasts transfected with control, Ctdnep1, and Eps8L2 siRNAs were microinjected with GFP-miniN2G to analyze TAN lines formation. Cells were stained for GFP (green) and phalloidin (magenta). TAN lines can be visualized in the control (yellow arrows). Line scans for each channel are represented in the right plots. Scale bars 10 μm.
(C) Quantification of the percentage of cells presenting at least one TAN line in wound-edge fibroblasts treated with control, Ctdnep1, and Eps8L2 siRNAs. The n value means number of experiments with >25 cells per experiment.
(D) Quantification of number of TAN lines per cell in the conditions described in (C). The n value means number of analyzed cells.
Data are represented as mean ± SEM. Statistics was performed by unpaired t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S3.