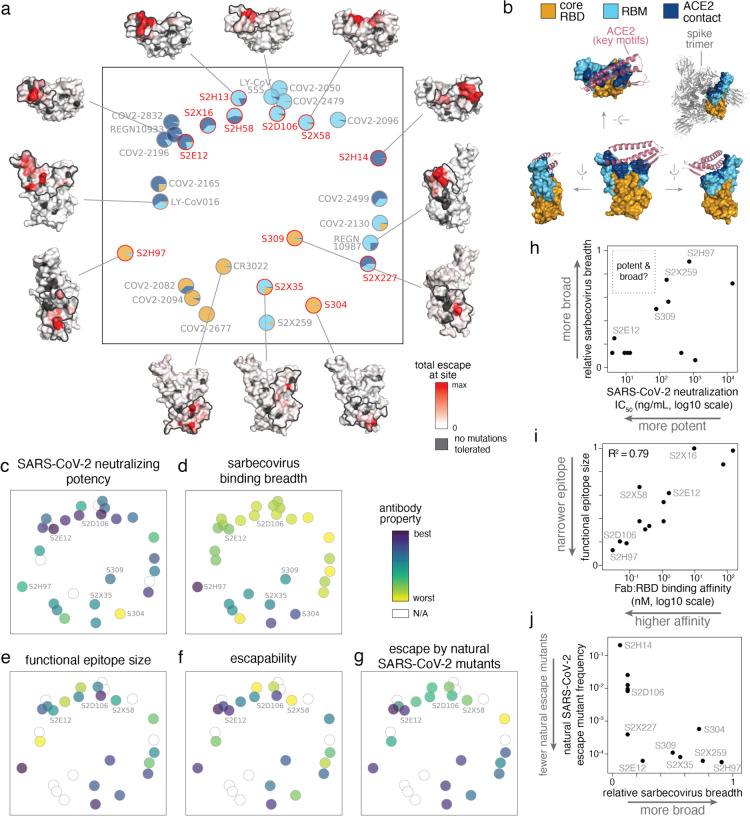
Fig. 2. Relationship between antibody epitope, potency, breadth, and escapability.
a, Multidimensional scaling projection of antibody epitopes based on similarities in sites of binding-escape as mapped in this (red) or prior (gray) studies. Pie charts illustrate the RBD sub-domains where mutations confer escape for each antibody [see key, (b)]. Structural projections of escape from representative antibodies are arrayed around the perimeter (scale bar, bottom right), with gray outlines tracing the structural footprint for antibodies with solved structures. b, Structural key for (a), illustrating the relative angles of structural views, the classification of sub-domains, and the context of ACE2 binding (only the interacting structural elements are shown) and spike quaternary structure. c-g, Projected epitope space from (a) annotated by antibody properties. For each property, antibodies are colored such that purple (scale bar, upper-right) reflects the most desirable antibody: most potent neutralization (IC50, log10 scale), highest breadth, narrowest functional epitope size, lowest escapability, and lowest natural frequency of escape mutants (log10 scale). See Methods for definition of each property, and Extended Data Fig. 6a for quantitative scales of each property and all of their pairwise correlations. h, Relationship between sarbecovirus breadth and SARS-CoV-2 neutralization potency for antibodies characterized in this study and S2X259 (see accompanying paper, Tortorici et al.). i, Relationship between functional epitope size and RBD binding affinity for antibodies characterized in this study and S2X259. Note that binding-escape selections were calibrated independently for each antibody (Extended Data Fig. 2, Methods), so this correlation is not a simple consequence of high-affinity antibodies having fewer mutations that reduce affinity below some global threshold of escape applied universally to all antibodies. j, Relationship between natural SARS-CoV-2 mutation escape (summed frequency of all binding-escape mutations on GISAID, Extended Data Fig. 4a) and sarbecovirus breadth for antibodies characterized in this study and S2X259.