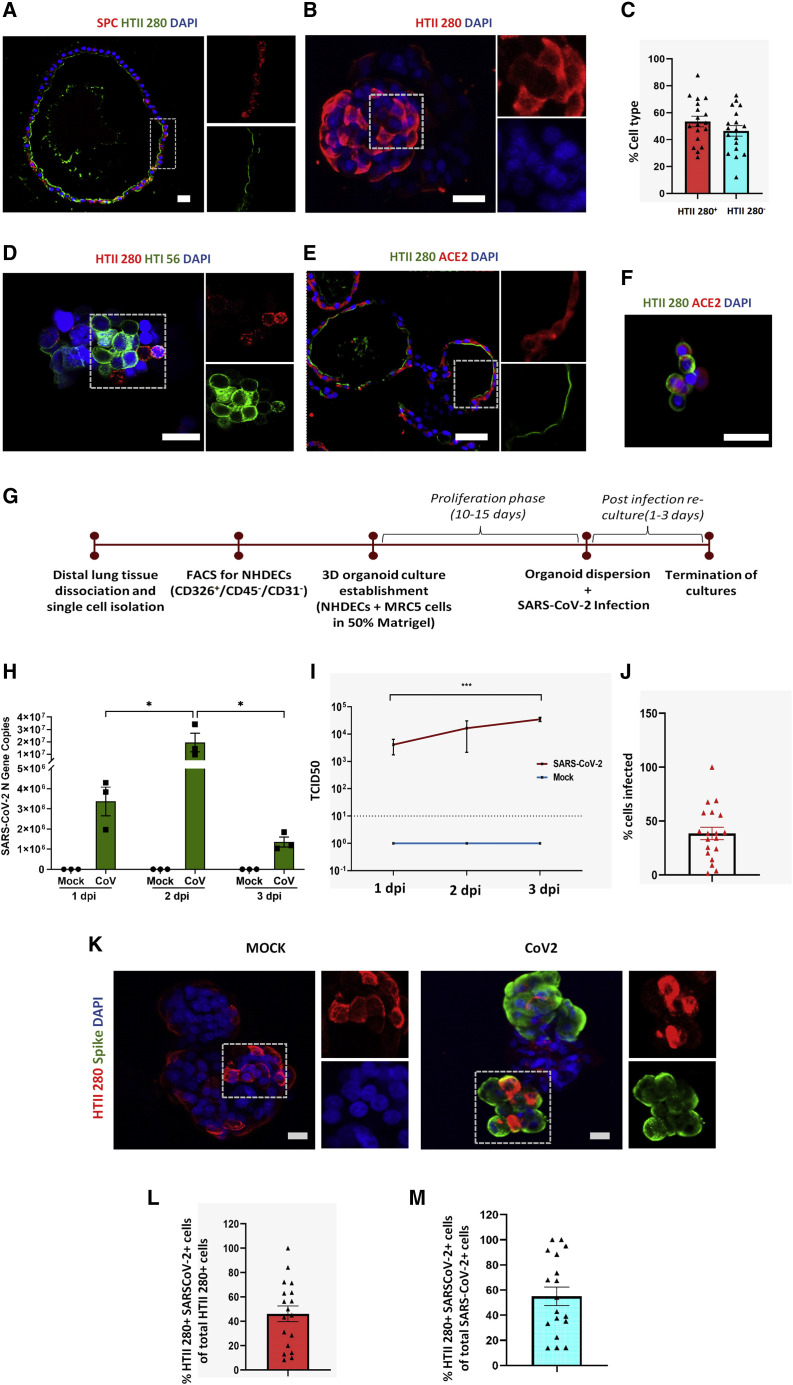
Figure 2.
SARS-CoV-2 infects normal human distal AT2 cells
(A) Sections of Matrigel-embedded alveospheres showing colocalization of AT2 markers HTII-280 and SPC.
(B) Whole-mount staining of alveospheres dispersed from Matrigel for AT2 marker HTII-280. Scale bar = 20 μm.
(C) Percentage of HTII-280-positive and HTII-280-negative cells in alveospheres in n = 3–5 fields from three biological replicates (two-tailed t test).
(D) Whole-mount alveospheres stained for the AT2 marker HTII-280 and AT1 cell marker HTI 56.
(E and F) ACE2 staining in sectioned alveospheres (E) and in AT2 cells (F) dissociated from alveospheres. Scale bar = 20 μm.
(G) Workflow for establishment of human distal alveolar cultures and their infection with SARS-CoV-2.
(H) Absolute N gene copy numbers in SARS-CoV-2-infected cultures peaked at 2 dpi; n = 3 independent cultures. Data were analyzed using two-way ANOVA with Sidak’s post hoc correction and represented as N gene copy number for individual cultures ± SEM. ∗p <0.05.
(I) Viral load in supernatant increased from 1 to 3 dpi. ∗∗∗p < 0.001.
(J) Percentage of total cells infected (n = 3).
(K) Infection was assessed at 2 dpi by antibody against SARS-CoV-2 “spike” protein (green) and colocalized with viral AT2 cell marker HTII-280 (red). Scale bar, 20 μm.
(L) Percentage of HTII-280-positive cells infected by SARS-CoV-2; n = 3–5 fields from each of three biological replicates.
(M) Percentage of total infected cells that are HTII-280-positive AT2 cells; n = 3–5 fields from each of three biological replicates.