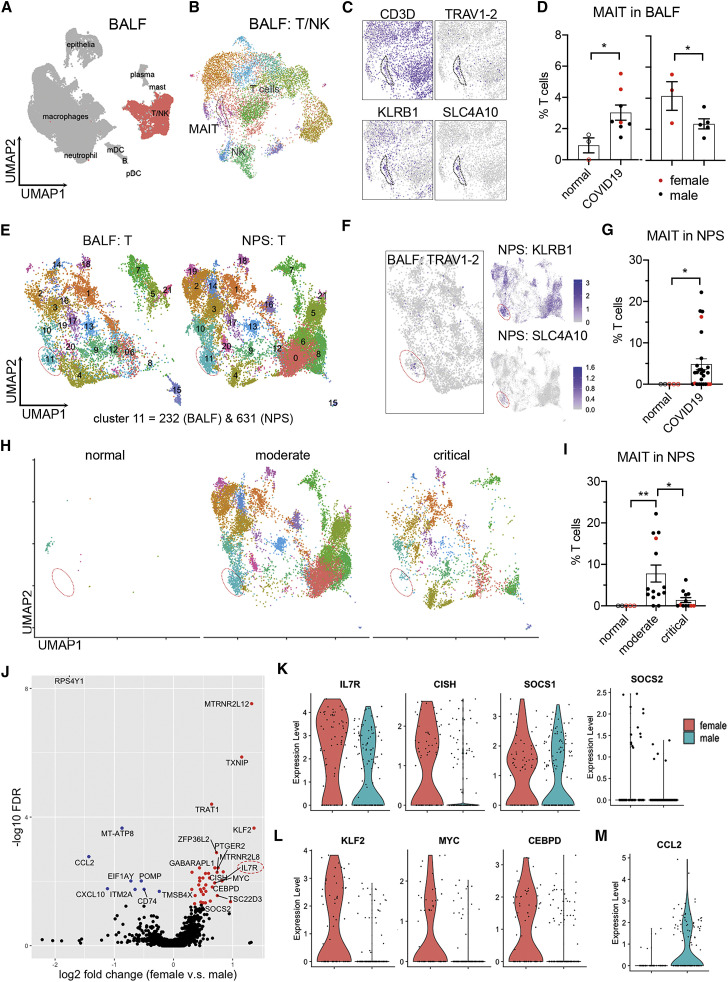
Figure 6.
Sex differences of MAIT cells in airway tissue samples from individuals with COVID-19
(A and B) Clustering analysis of scRNA-seq data from the COVID-19 BALF dataset with subtracted T and NK cells.37
(C) MAIT cell cluster indicated by marker genes.
(D) Frequencies of MAIT cells in BALF between normal subjects and individuals with COVID-19 (left) and across sexes within subjects with COVID-19 (right).
(E) Integrated clustering analysis of NPSs with BALF using Seurat 3. Cluster 11, MAIT cells.
(F) Referenced MAIT cell cluster in NPSs by expression of TRAV1-2 in BALF and the indicated marker genes in NPSs.
(G) Frequencies of MAIT cells in NPSs from healthy subjects and subjects with COVID-19.
(H and I) Visualization (H) and frequencies (I) of MAIT cells in NPSs, grouped by disease severity.
(J) Volcano plot showing DEGs of BALF MAIT cells between sex with log2 fold change and −log10 FDR.
(K–M) Expression of DEGs in IL-7 signaling (K), transcription factors (L), and CCL2 (M).
Data were plotted as mean ± standard error (D, G, and I), with females in red and males in black. Significance was determined by unpaired one-tailed Student’s t test (D), Mann-Whitney test (G), and Kruskal-Wallis test with Dunn’s post hoc test (I): ∗p < 0.05, ∗∗p < 0.01. See also Figure S5.