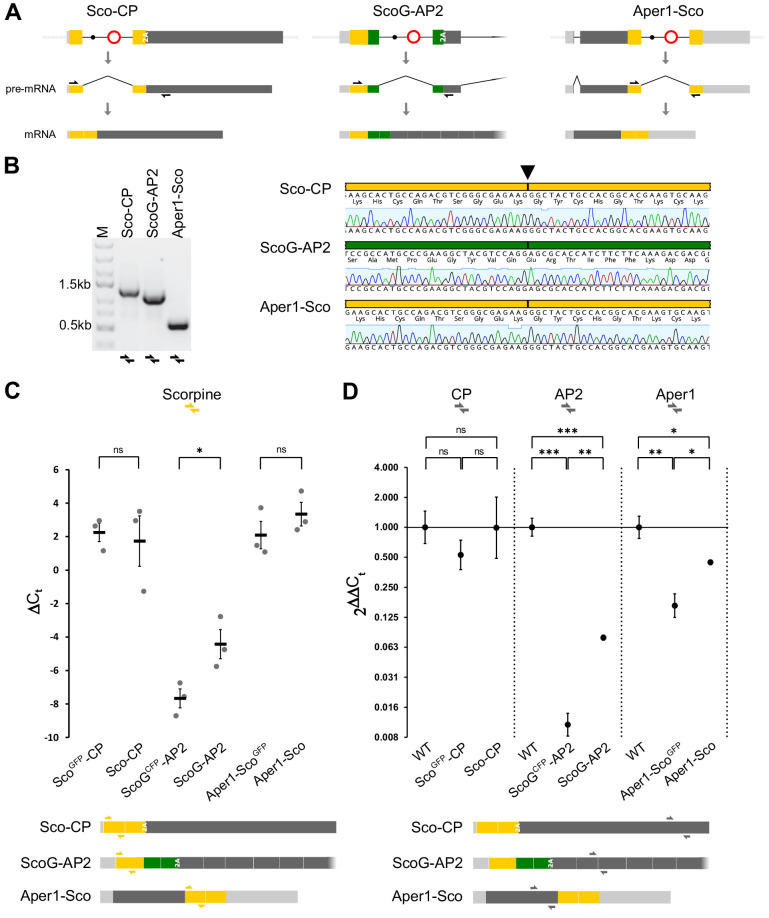
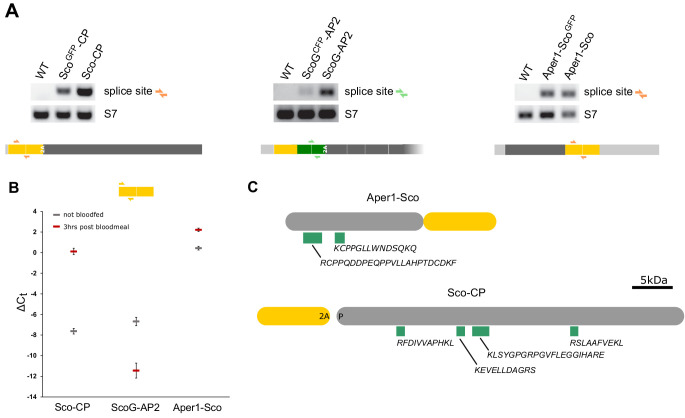
Figure 3. Splicing of the artificial intron and expression analysis.
(A) Schematic showing mRNA expression of the modified host genes of the three markerless strains assuming correct splicing of the artificial intron. Black arrows indicate RT-PCR primers used in B. (B) RT-PCR (left) and sequencing of the amplicons (right) indicate precise splicing of the three artificial introns. Midguts of homozygous strains Sco-CP, ScoG-AP2, and Aper1-Sco were dissected 3 hr after blood feeding and RNA was extracted for RT-PCR. (C) Relative mRNA expression of Scorpine in the transgenic strains with and without the marker cassette. (D) Expression of the host genes CP, AP2, and Aper1 in transgenic strains with and without the marker cassette relative to the wild type. RNA was extracted from 10 to 15 midguts 3 hr after the blood meal in the case of CP and Aper1, and non-blood-fed for AP2. qPCR with the primer pairs indicated as coloured half arrows in the schematic below was conducted on cDNA and expression was normalized to the S7 house-keeping reference gene. Data derive from three biological replicates with three technical replicates each. p-values were calculated on ΔCt values using the unpaired Student’s t-test. *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, and ns: not significant.