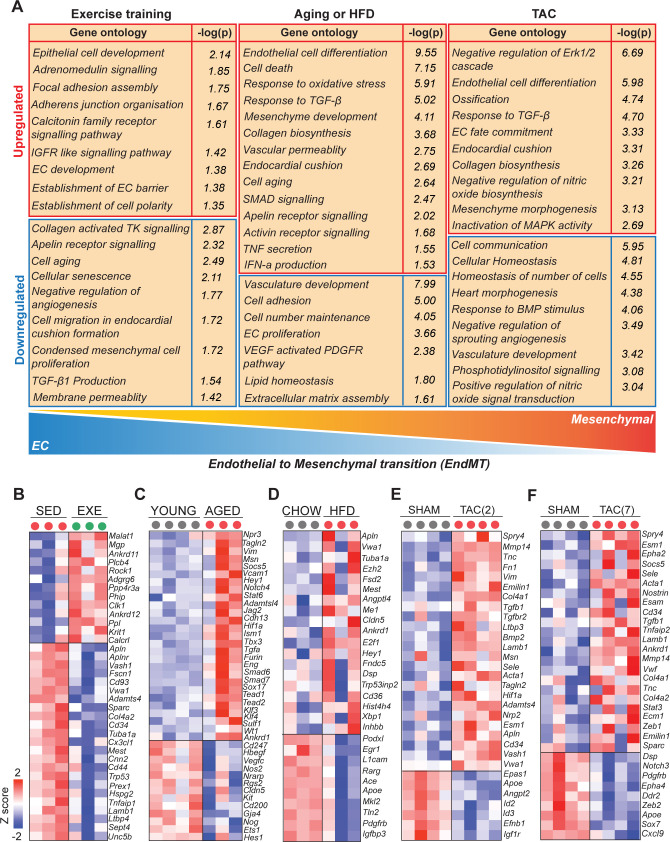
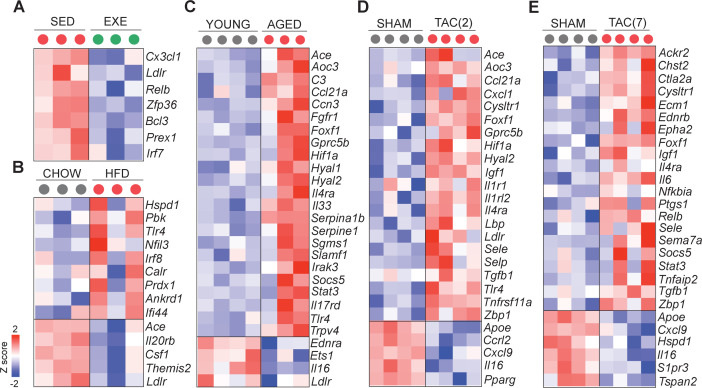
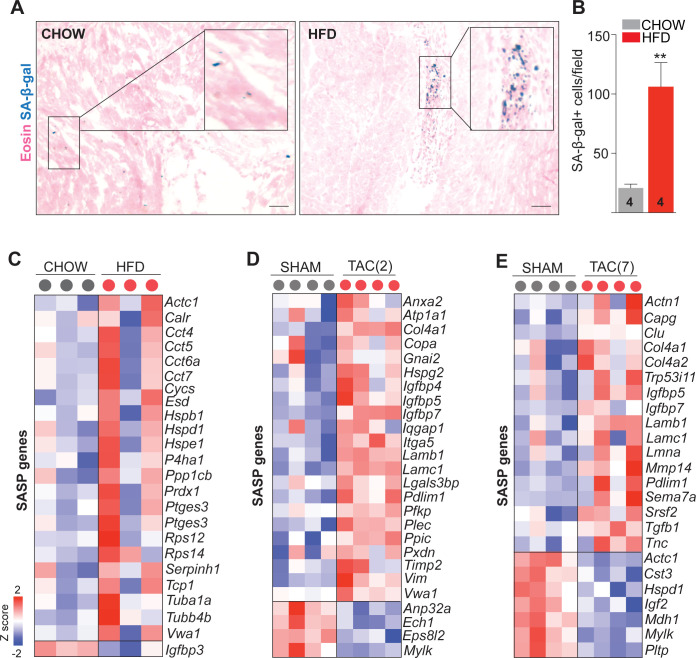
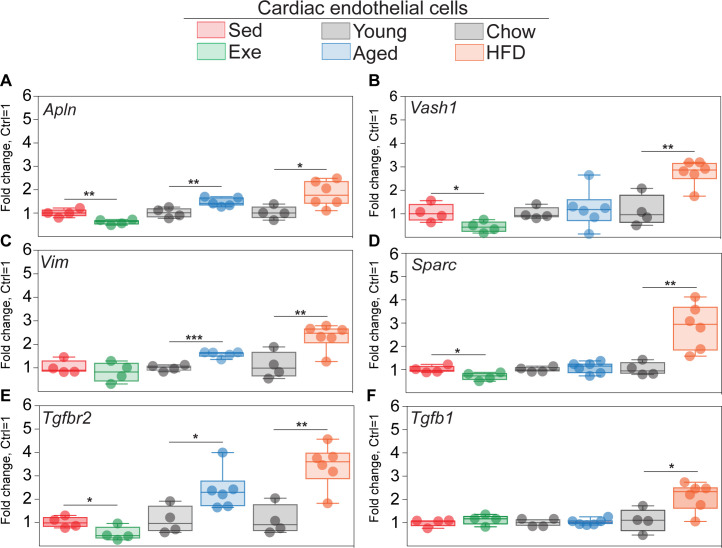
Figure 3. Cardiovascular disease (CVD) risk factors activate mesenchymal gene expression in cardiac endothelial cells (ECs).
(A) Gene ontology analysis of the up- and downregulated genes. Note the opposite changes induced by exercise training compared to the CVD risk factors. (B–F) Heatmaps showing the differential gene expression of endothelial and mesenchymal genes previously associated with endothelial-to-mesenchymal transition (EndMT). Genes are selected based on published data sets (references are found in Figure 3—source data 1). In all panels, the up- and downregulated genes with the false discovery rate (FDR; Benjamini–Hochberg adjusted p-value) threshold of 0.05 were considered. In the heatmap, each color-coded circle (red, green, and black) indicates an individual biological sample within each experimental group. N = 3–4 male mice/group.