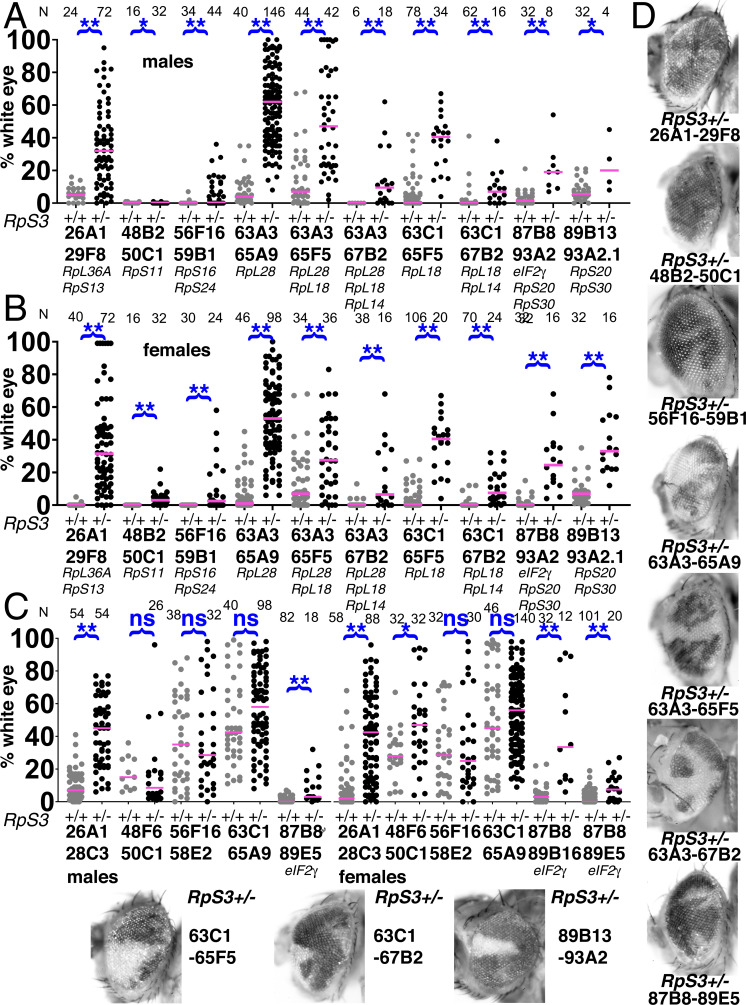
Figure 9. Elimination of segmentally aneuploid clones requires a Rp+/+ background.
Eye contributions of clones of segmentally aneuploid genotypes deleting the indicated Rp loci. in RpS3+/+ and RpS3+/- genetic backgrounds. Magenta bars show the median contributions, N as indicated for each genotype. (A) In males, the RpS3+/- genetic background (black datapoints) allows for significantly greater contribution in all cases except Df(48B2-50C1)/+. No Df(3R)89B13-93A2.2/RpS3 flies were obtained, this genotype is lethal due to an unidentified shared lethal outside the 89B13-93A2.2 region. (B) Comparable data from females. The RpS3+/- genetic background (black datapoints) always allows for significantly greater contribution. (C) Eye contributions of clones of segmentally aneuploid cells where no Rp loci are affected. The RpS3+/- genetic background (black datapoints) had no significant effect on many such genotypes, but did enhance the contribution of Df(26A1-28C3)/+ clones and of the Df(3R)87B8-89B16/+ and Df(3R)87B8-89E5/+ clones that were haploinsuffiicent for eIF2γ +. No Df(3R)87B8-89B16/RpS3 males were obtained. (D) Representative examples of these RpS3+/- genotypes (males shown). The RpS3+/+ control data were shown previously in Figure 2, except for 87B8-89B16 females. Statistics. Pairwise comparisons using the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). ns – difference not statistically significant (p>0.05). * - difference significant (p<0.05). ** - difference highly significant (p<0.01). Genotypes: Same as for Figure 3, with an FRT82B RpS3 chromosome substituting for FRT82B where indicated.