Fig. 4. TEV13 and FphF16 are potent and specific covalent inhibitors.
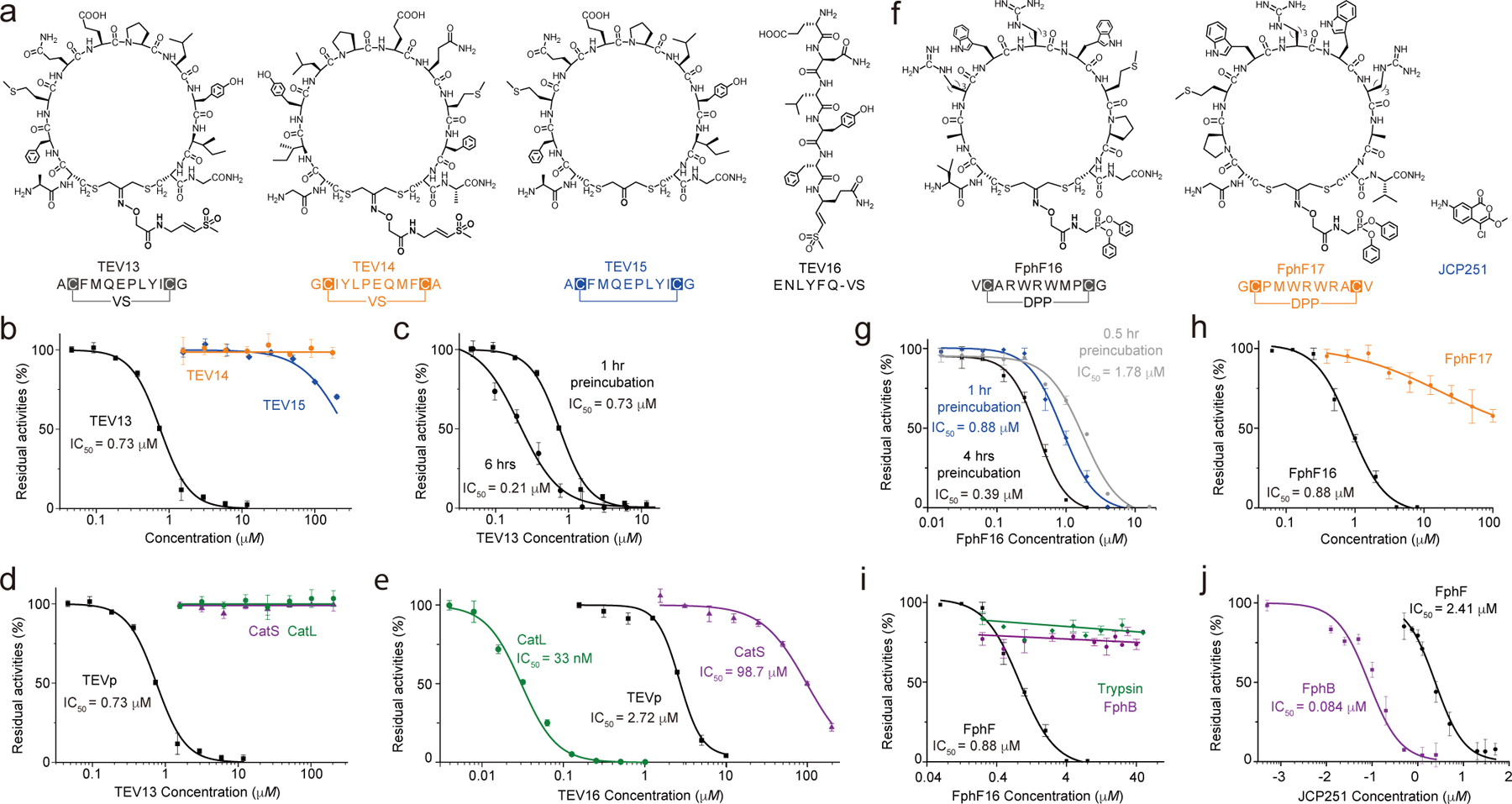
a) Structures of the optimized cpVS inhibitor TEV13 compared to the scrambled isomer, TEV14, the cyclic peptide lacking the VS electrophile in the linker, TEV15, and the linear peptide VS synthesized based on the conserved substrate specificity of TEV. b) Dose response inhibition studies of the cyclized peptides using recombinant TEV. Plots show residual enzyme activity over a range of inhibitor doses as measured by hydrolysis of a fluorogenic substrate. Enzyme was pre-treated with inhibitor at the indicated concentrations for 1 hour followed by addition of substrate and measurement of activity. IC50 values, when measurable are indicated. c) Time dependence of inhibition of TEV protease by TEV13. Recombinant enzyme was preincubated with a range of TEV13 concentrations for 1 hr or 6 hrs as indicated followed by measurement of residual activity using a fluorogenic substrate. Comparisons of the activity of (d) TEV13 and (e) TEV16 for TEV and the off-target cysteine proteases Cat S and Cat L. Inhibition studies were performed as in (b). (f) Structure of the optimized cpDPP inhibitor FphF16 compared to the sequence scrambled cpDPP, FphF17, and JCP251, a reported chloroisocoumarin inhibitor of the Fph hydrolases. g) Time dependence of inhibition of FphF hydrolase by FphF16. FphF16 was pre-incubated with the recombinant FphF hydrolase for 0.5 hr, 1 hr and 4 hrs, followed by the addition of a fluorogenic substrate, 4-methylumbelliferyl heptanoate. The residual enzyme activities represented by the fluorescent singles were plotted. h) Dose response of FphF16 and FphF17 to inhibit recombinant FphF hydrolase. IC50 values were determined as above. Comparisons of the activity of (i) FphF16 and (j) FphF17 for FphF hydrolase and the off-target FphB hydrolase and trypsin. Inhibition studies were performed as in (g). n= 3 biologically independent wells (mean + s.d. is depicted).
