Fig. 1 |. MPL accurately recovers selection from complex dynamics.
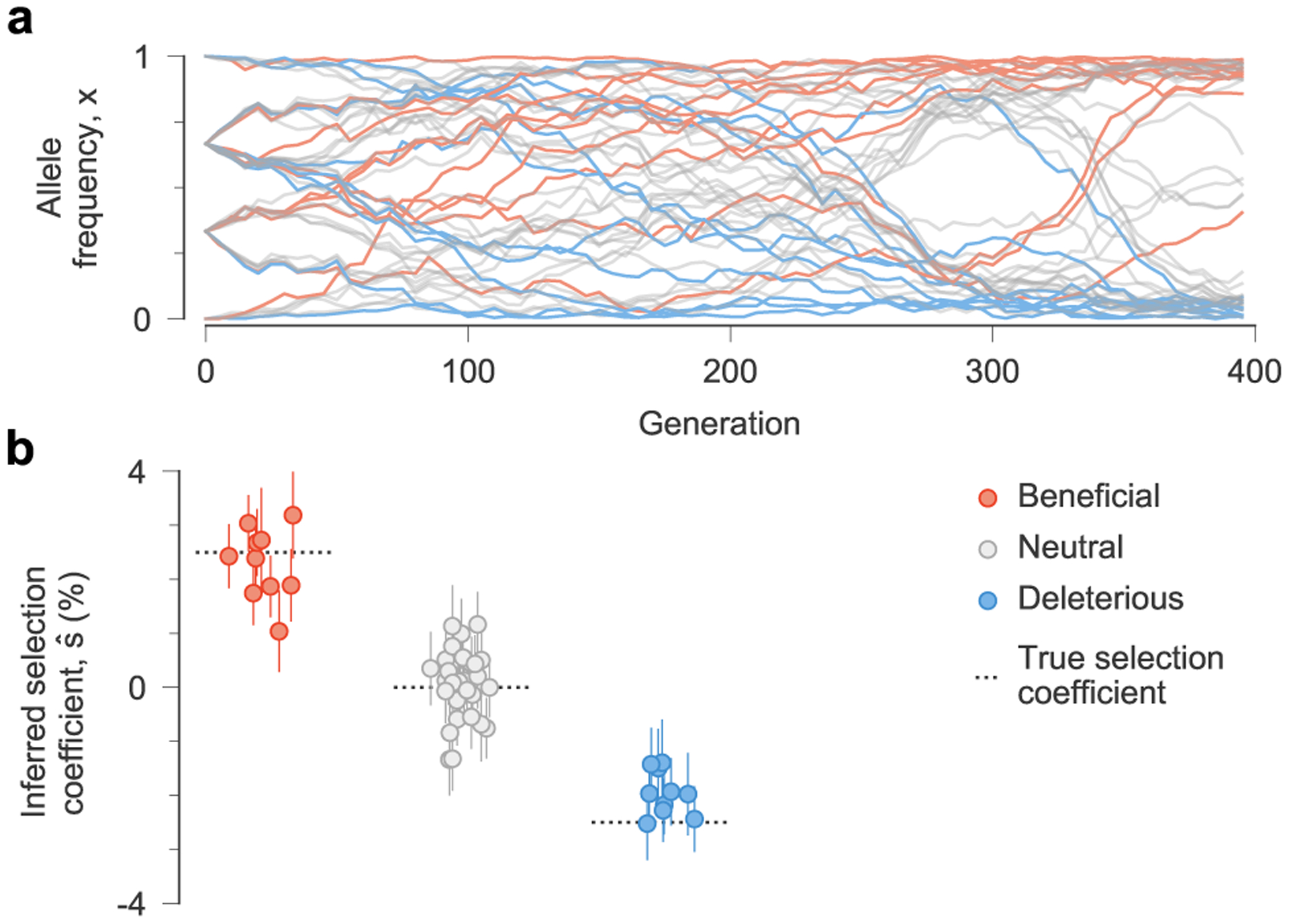
a, Simulated allele frequency trajectories in a model with ten beneficial, 30 neutral and ten deleterious mutant alleles. The initial population is a mix of three subpopulations with random mutations. Selection is challenging to discern from individual trajectories alone. b, Selection coefficients inferred by MPL, presented as mean values ± 1 theoretical s.d. (Methods), are close to their true values. Simulation parameters. L = 50 loci with two alleles at each locus (mutant and WT): ten beneficial mutants with s = 0.025, 30 neutral mutants with s = 0 and ten deleterious mutants with s = −0.025. Mutation probability per locus per generation μ = 10−3, population size N = 103. The initial population is composed of approximately equal numbers of three random founder sequences, evolved over T = 400 generations.
