Extended Data Fig. 7 |. For most variants, effects on inferred selection coefficients for other variants, and linkage disequilibrium, are stronger at smaller genomic distances.
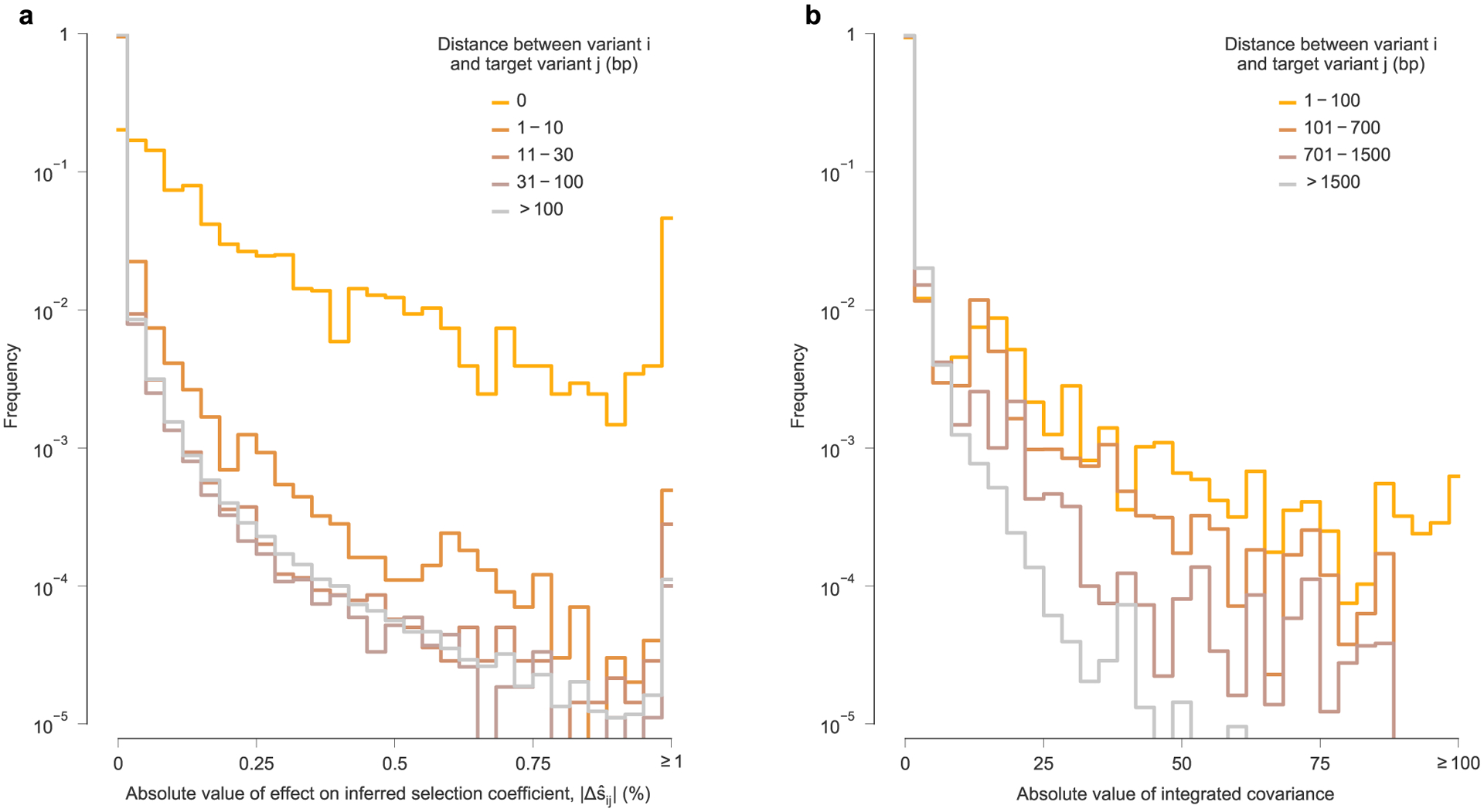
a, Histogram of the absolute value of linkage effects on inferred selection coefficients for other variants , divided into subgroups based on the distance along the genome between variant i and target variant j. Consistent with intuition, the large effects on inferred selection coefficients occur most frequently for different variants that occur at the same site on the genome (that is, distance equal to zero). ‘Interactions’ between such variants are necessarily perfectly competitive because only a single nucleotide is allowed at each position in the genetic sequence. For most variants, stronger linkage effects on inferred selection coefficients are more frequently observed for other variants within a distance of ten base pairs (bp). Large linkage effects for pairs of variants within a distance of 30 bp, the approximate length of a linear T cell epitope, occur appreciably more frequently than for pairs of variants at greater genomic distances. However, there is little difference in the distribution of linkage effect sizes for pairs of variants that are between 31 bp and 100 bp apart compared to pairs of variants that are more than 100 bp apart. Nonetheless, some strong linkage effects on inferred selection are observed at long genomic distances (see Fig. 4 and Supplementary Fig. 5). b, Linkage disequilibrium, measured by the absolute value of the off-diagonal entries of the integrated allele frequency covariance matrix, Cint. Like the , linkage decays along with the distance between variants along the genome. However, we note that linkage disequilibrium values in general appear to be more long-ranged.
