Extended Data Fig. 2 |. MPL improves selection inference for simulated data sets.
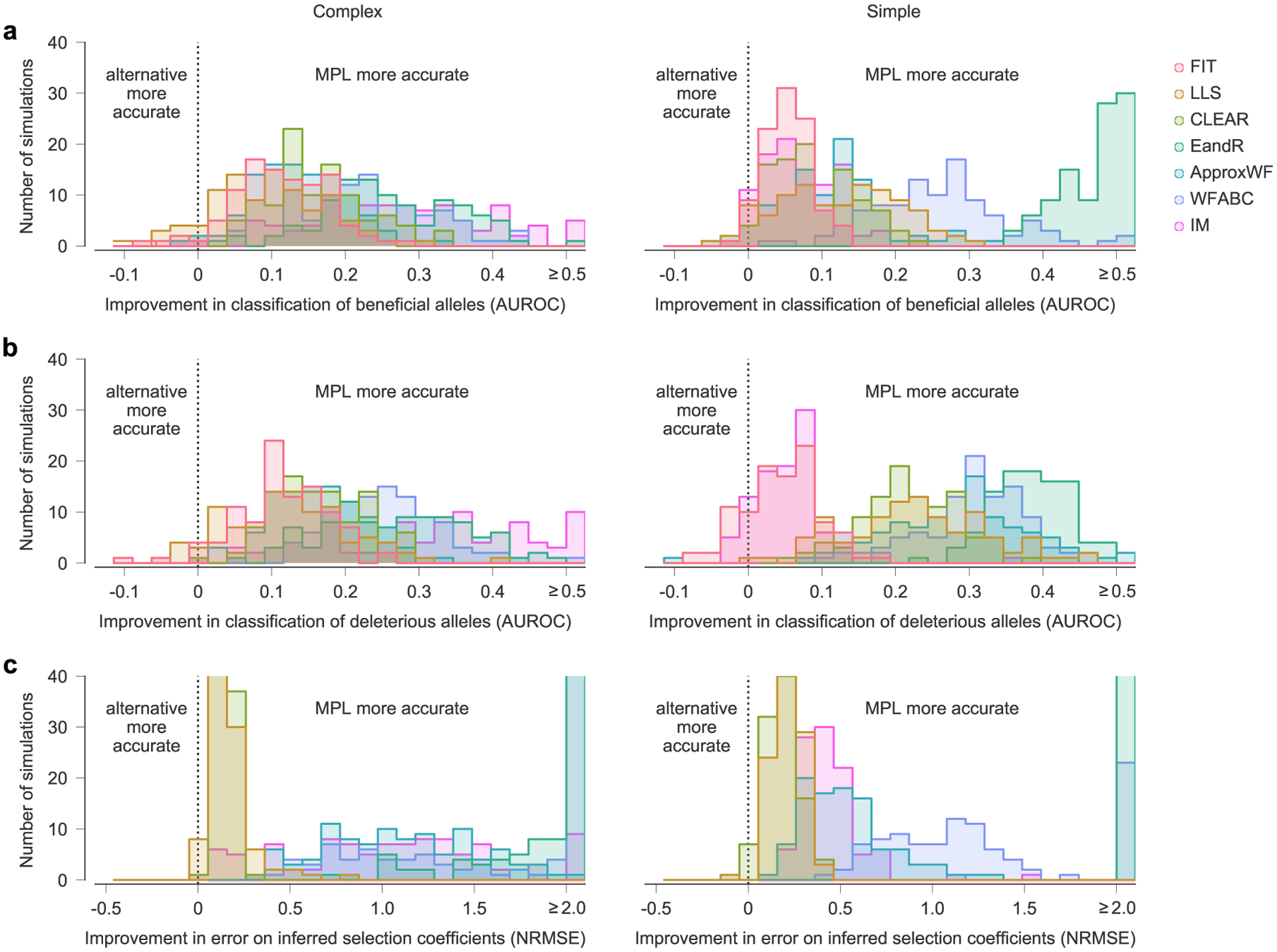
In Fig. 2, we showed the performance of MPL and existing methods on simulated test data, averaged over n = 100 replicate simulations with identical parameters. Here we show the improvement of MPL over existing methods for the classification of beneficial (a) and deleterious (b) mutations, and for the error in the estimated selection coefficients (c), for each individual simulation. Selection is more difficult to infer in some simulated data sets, but results from MPL show better agreement with the true parameters in the vast majority of simulations. Simulation parameters. L = 50 loci with two alleles at each locus (mutant and WT): ten beneficial mutants (with s = 0.1 for complex, s = 0.025 for simple), 30 neutral mutants (s = 0 for both scenarios), and ten deleterious mutants (s = −0.1 for complex, s = −0.025 for simple). Mutation probability μ = 10−4, population size N = 103. For the complex case, the initial population is composed of equal numbers of five random founder sequences, evolved over T = 310 generations. Recorded trajectory used for inference begins at generation 10. For the simple case, the initial population begins with all WT sequences, evolved over T = 1000 generations.
