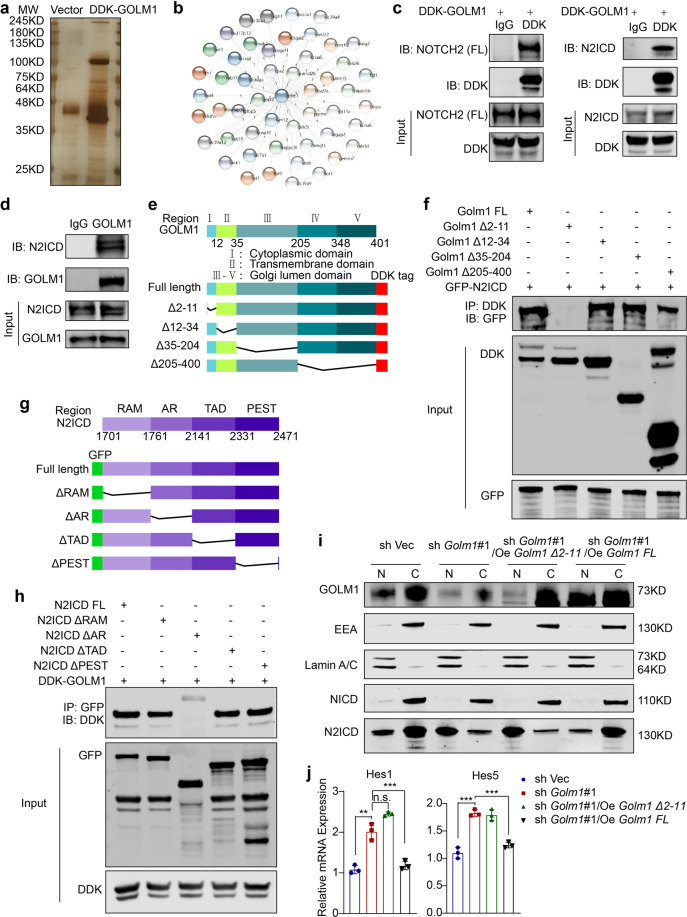
Fig. 6.
GOLM1 interacts with N2ICD and modulates its downstream signaling. a Mass spectrometry (MS) analysis of GOLM1-associated proteins. Total cell lysates from DDK-GOLM1 expressed cells were subjected to affinity purification. The purified protein complex was resolved on SDS-PAGE and silver stained; then the bands were retrieved and analyzed by MS. b A diagram depicts GOLM1 interactors as detected by MS (Detailed information in Supplementary Information). c The interaction between GOLM1 and NOTCH2/N2ICD in 293T cells stably expressing DDK GOLM1 was detected by immunoprecipitation. d The interaction between GOLM1 and NOTCH2 (FL)/N2ICD in Caco-2 cells was detected by immunoprecipitation. e Diagrammatic representation of GOLM1 and its truncated forms. Based on sequence and structure analyses, cytoplasmic domain, transmembrane domain, and Golgi lumen domain are indicated. f Various GOLM1 truncation constructs tagged with DDK were co-transfected with GFP-NOTCH2 in 293T cells for domain mapping. Immunoprecipitation analysis was performed with anti-GFP or anti-DDK antibodies. g Diagrammatic representation of N2ICD and its truncated forms. h Various N2ICD truncation constructs tagged with GFP were co-transfected with DDK-GOLM1 in 293T cells for domain mapping. Immunoprecipitation analysis was performed with anti-GFP or anti-DDK antibodies. i Cellular fractionations from GOLM1-deficient Caco-2 cells transfected with various GOLM1 truncation constructs and control cells were analyzed by immunoblotting with the indicated antibodies. j The relative mRNA expression levels of Notch signaling downstream genes from GOLM1-deficient Caco-2 cells transfected with various GOLM1 truncation constructs and control cells were determined by qRT-PCR (the data are represented as the means ± SEM; **P < 0.01, ***P < 0.001; unpaired, two-tailed Student’s t test)