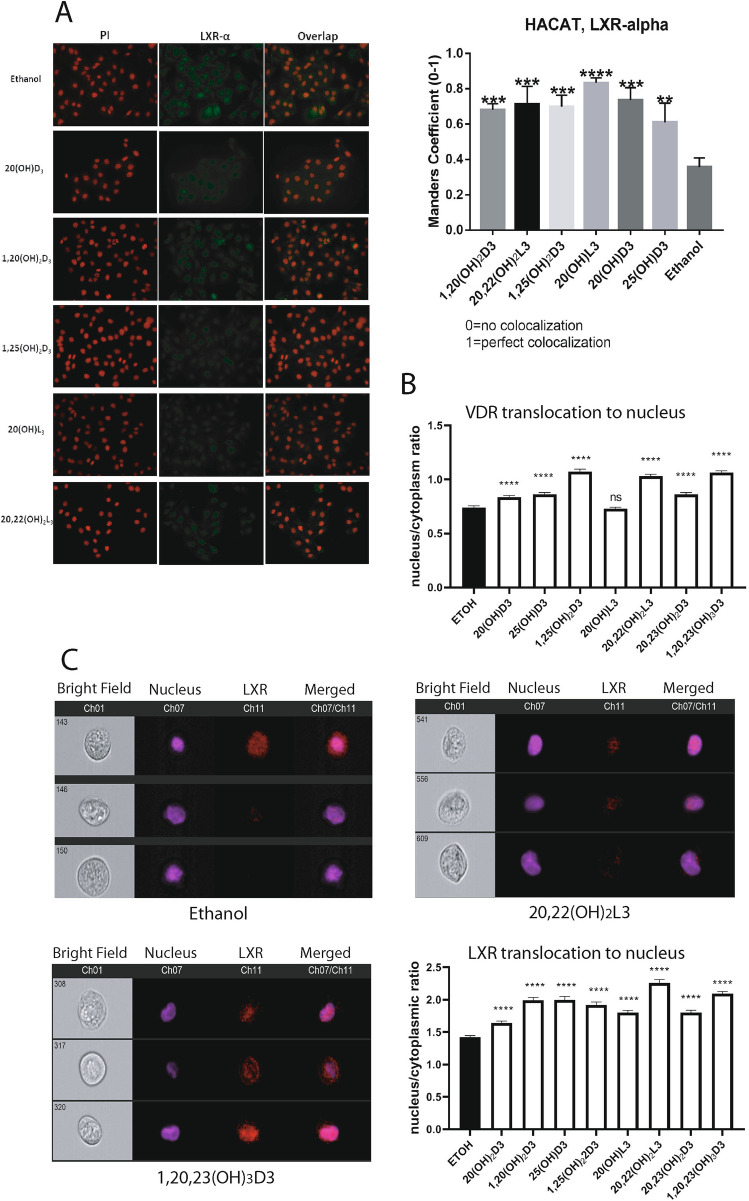
Figure 6.
Ligand induced translocation of LXR to the nucleus. (A) Colocalization analysis of LXRα and PI (nuclear counterstain) in HaCaT cells treated with 10–7 M of 25(OH)D3, 1,25(OH)2D3, 20(OH)D3, 1,20(OH)2D3, 20(OH)L3 or 20,22(OH)2L3 or ethanol (control) for 24 h. Manders’ coefficient (0–1) (right panel) was significantly higher for cells treated with D3 and L3-hydroxyderivatives than cells treated with vehicle only. Data are presented as means ± SD, (n = 2). (B) Imaging flow cytometry analysis of HaCaT cells treated with ethanol or 10–7 M 20(OH)D3, 1,20(OH)2D3, 25(OH)D3, 1,25(OH)2D3, 20(OH)L3, 20,22(OH)2L3, 20,23(OH)2L3 or 1,20,23(OH)3D3 for 12 h. Fixed cells were stained with Hoechst and immunostained with antibodies against VDRR. Ratios of nuclear (co-localization with Hoechst) vs cytoplasmic localization of VDR were determined following analysis of 515 to 2339 individual cells. (C) Imaging cytometry images of individual HaCaT cells showing LXR localization in cytoplasm or nucleus following treatment with ethanol, 20,22(OH)2L3 or 1,20,23(OH)3D3. Bar graphs represent quantitative analysis of images acquired by imaging cytometry. HaCaT cells treated with ethanol or 10–7 M 20(OH)D3, 1,20(OH)2D3, 25(OH)D3, 1,25(OH)2D3, 20(OH)L3, 20,22(OH)2L3, 20,23(OH)2L3 or 1,20,23(OH)3D3 for 12 h were fixed, permeabilized cells and immunostained with Hoechst and antibodies against LXR. Ratios of nuclear vs cytoplasmic localization of LXR were determined following analysis of 515–2339 individual cells. The data in bar graphs (A–C) show significant differences between ligand -treated and control (ethanol treated) cells. Analysis was done using t-test: **p < 0.01, ***p < 0.001 or ****p < 0.0001 versus control (ethanol). For part A the slides were examined using a KEYENCE America BZ-X710 Fluorescence Microscope (Itasca, IL) and captured using KEYENCR BZ-X viewer (version 1.3.0.5, https://www.keyence.com/products/microscope/fluorescence-microscope/bz-x700/index_pr.jsp). The images were subsequently analyzed using the JACoP plugin (version 2.1.1, https://imagejdocu.tudor.lu/doku.php?id=plugin:analysis:jacop_2.0:just_another_colocalization_plugin:start) for colocalization analysis1 with ImageJ (version 1.52a, http://imageJ.nih.gov/ij). For part C, images were captured using an Amnis ImageStreamX Mk II Imaging Flow Cytometer (Luminex Corporation) and IDEAS software version 6.2.