Figure 2.
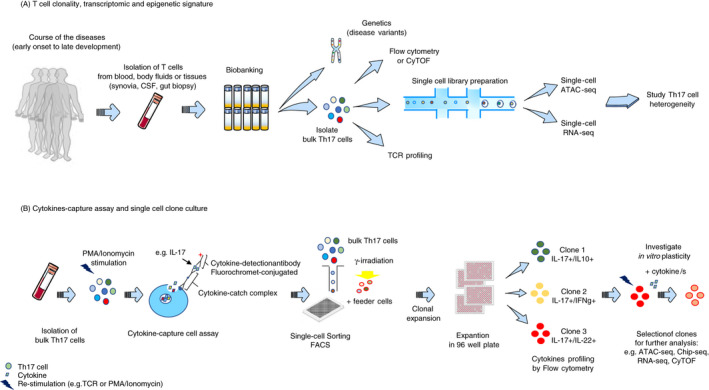
Current methods to study human Th17 cell heterogeneity and plasticity. A) To resolve the heterogeneity and dynamic of specific Th17 populations in a disease setting, it is relevant to isolate T cells from blood, other body fluids or tissues (e.g. synovia, cerebrospinal fluid (CSF), gut biopsy) at several time‐points (e.g. before and after treatment, at multiple disease stages). Collected cells can be biobanked for traceability and used at later time‐points. Genetic analysis can be performed to identify risk variants associated with the disease and altered Th17 cell phenotype and function, and, in parallel, perform TCRVβ sequencing to investigate clonality. Flow cytometry (FACS) or mass cytometry (CyTOF) allows to immunophenotype bulk Th17 cell populations to dissect their complexity. Th17 cell heterogeneity can be resolved by ex vivo epigenetic and transcriptional deep phenotyping at single‐cell level combining, for example, ATAC‐seq and RNA‐seq. Isolation of cytokine‐specific Th17 cells by cytokine capture assay allows in vitro generation of homogeneous Th17 clones. Isolation of bulk Th17 cells from, for example, blood and their stimulation with PMA/ionomycin allow single‐cell sorting, by FACS, of specific cytokine‐expressing Th17 cells and culturing with feeder cells. Following clonal expansion, each clone is transferred and expanded in a 96‐well plate. Th17 clones can be immunophenotyped before and after restimulation, which allows clone selection for further investigation. Plasticity of a defined Th17 clone can be investigated after reactivation and culturing with cytokines using CyTOF, RNA‐seq, ATAC‐seq and Chip‐seq.
