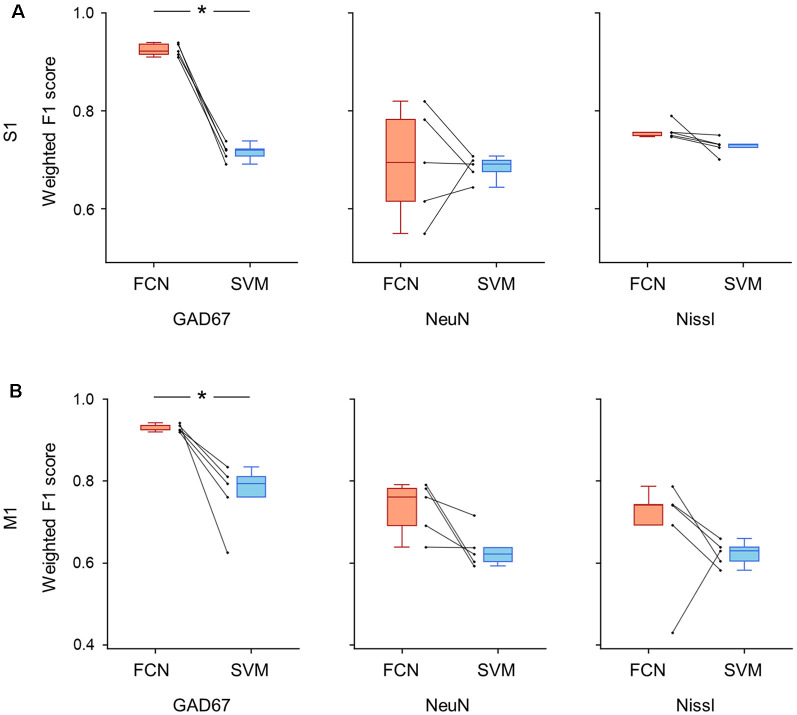
Figure 8.
Classification performance of cell images using single fluorescent signals. (A) Weighted F1 score of the FCN model (red) and the PCA-SVM classifier (blue) trained on cell images from S1. Left: classification performance using an anti-GAD67 signal. Each point (gray) signifies the score for the cross-validation (5-fold). P = 5.6 × 10–5, t(4) = 18.0, n = 5-fold cross-validations, paired t-test. *P < 0.05. Middle, the same as on the left, but for an anti-NeuN signal. P = 0.87, t(4) = 0.18, n = 5-fold cross-validations, paired t-test. Right: the same as on the left, but for a Nissl signal. P = 0.09, t(4) = 2.18, n = 5-fold cross-validations, paired t-test. (B) The same as (A), but for M1. Left, P = 0.01, t(4) = 4.1, n = 5-fold cross-validations, paired t-test. *P < 0.05. Middle, P = 0.06, t(4) = 2.6, n = 5-fold cross-validations, paired t-test. Right, P = 0.22, t(4) = 1.45, n = 5-fold cross-validations, paired t-test. Abbreviations: S1, primary somatosensory cortex; M1, primary motor cortex; FCN, fully convolutional network; SVM, support vector machine; PCA, principal component analysis.