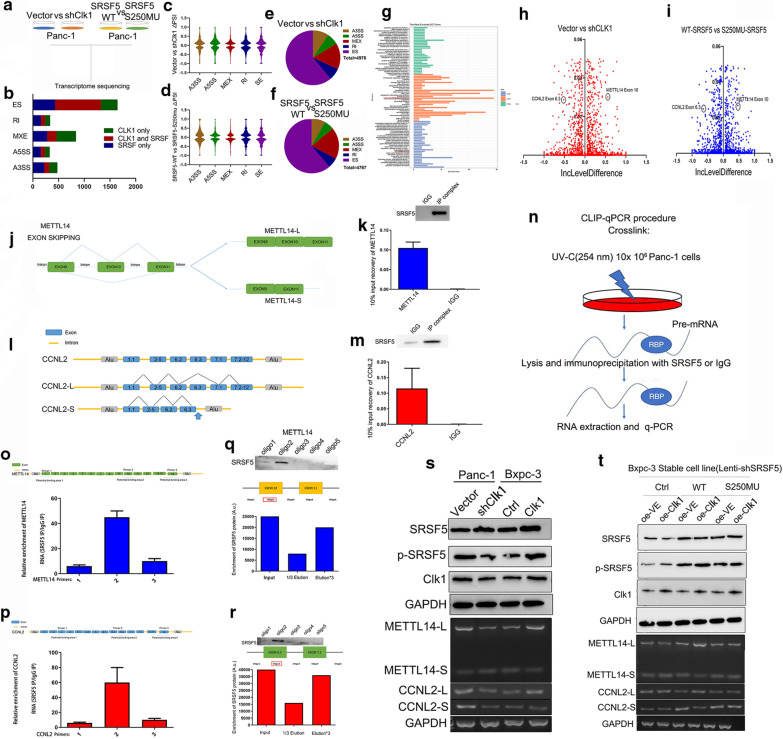
Fig. 6.
SRSF5 controlled the exon skipping of METTL14 and CyclinL2. a A schematic diagram shows the experimental design of transcriptome sequencing in Group A and Group B. b Genes with different AS patterns were represented according to the type of AS events. c, d The constituent ratios of different AS events were showed in two groups. e, f The changes of PSI of different AS events were showed in two groups. g GO analysis of genes with shift of exon skipping in two groups. h, i Analysis of △PSI in two groups: X axis represents IncLevelDifference: Difference expression of Exon Inclusion Isoform between Vector and sh-CLK1 (h) or overexpressed SRSF5wt and SRSF5mu (i). Y axis represents FDR. j, l Schematic diagrams show the alternative splicing of METLL14 (J) and Cyclin L2 (L) by exon skipping. k, m RIP assays were performed to confirmed the interaction between SRSF5 and pre-mRNA of METTL14 (k) or CyclinL2 (m). n Schematic diagrams show the CLIP-qPCR. CLIP assays were performed in Panc-1 cells to confirmed the interaction between SRSF5 and the region of METTL14 (o) or CyclinL2 (p). q, r A series of bait-oligo were constructed, and RNA pull down assays were conducted to identified the site of METTL14 (q), or CyclinL2 (r) that SRSF5 interacted with in PANC-1 cells. s, t Representative images and quantification of the relationships between CLK1-SRSF5 axis and AS patterns of METTL14 (s) and Cyclin L2 (t) are presented. n = 3 for each group; data are shown as mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, between the indicated groups