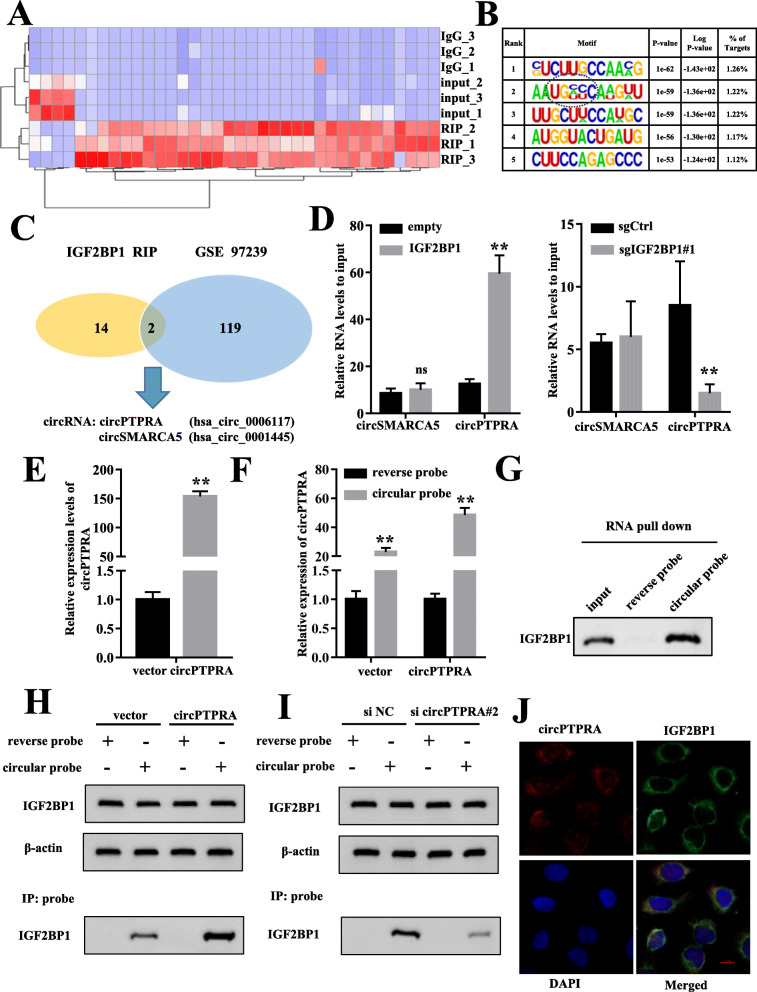
Fig. 2.
IGF2BP1 could interact with circPTPRA in the cytoplasm in BC cells. a Heatmap of the RIP-seq experiments showing the IGF2BP1-binding circRNAs in bladder cancer (BC). b Top consensus sequences of IGF2BP1-binding sites and the m6A motif detected by HOMER motif analysis. c Overlap of IGF2BP1-binding circRNAs identified by RIP–seq and published GSE 97239 dataset in BC. d RIP and qRT-PCR assays using an antibody specific for IGF2BP1 showing the interaction between circRNAs (circPTPRA, circSMARCA5) and IGF2BP1 in T24T cells with stable overexpression or knockout of IGF2BP1. e qRT-PCR analysis verified the efficiency of circPTPRA overexpression and pulled down by biotin-labeled reverse or circular RNA probes from lysates of T24T cells. f Cell lysates prepared from T24T cells transfected with circPTPRA or the vector were hybridized with circular probe or reverse probe for RNA pull-down assays. g Western blot assays showing the IGF2BP1 protein pulled down by biotin-labeled reverse or circular RNA probes from lysates of T24T cells. h Cell lysates prepared from T24T cells transfected with circPTPRA or the vector were subject to immunoprecipitation with IGF2BP1 and were also hybridized with reverse or circular RNA probes for IP assays with anti-IGF2BP1 antibody, followed by western blot. i Cell lysates prepared from T24T cells transfected with si-circPTPRA or the mock were subject to immunoprecipitation with IGF2BP1 and were also hybridized with reverse or circular RNA probes for IP assays with anti-IGF2BP1 antibody, followed by western blot. j Dual RNA-FISH and immunofluorescence staining assay indicating the colocalization of circPTPRA (red) and IGF2BP1 (green) in T24T cells (scale bar represents 10 μm). **, P < 0.01