Figure 5:
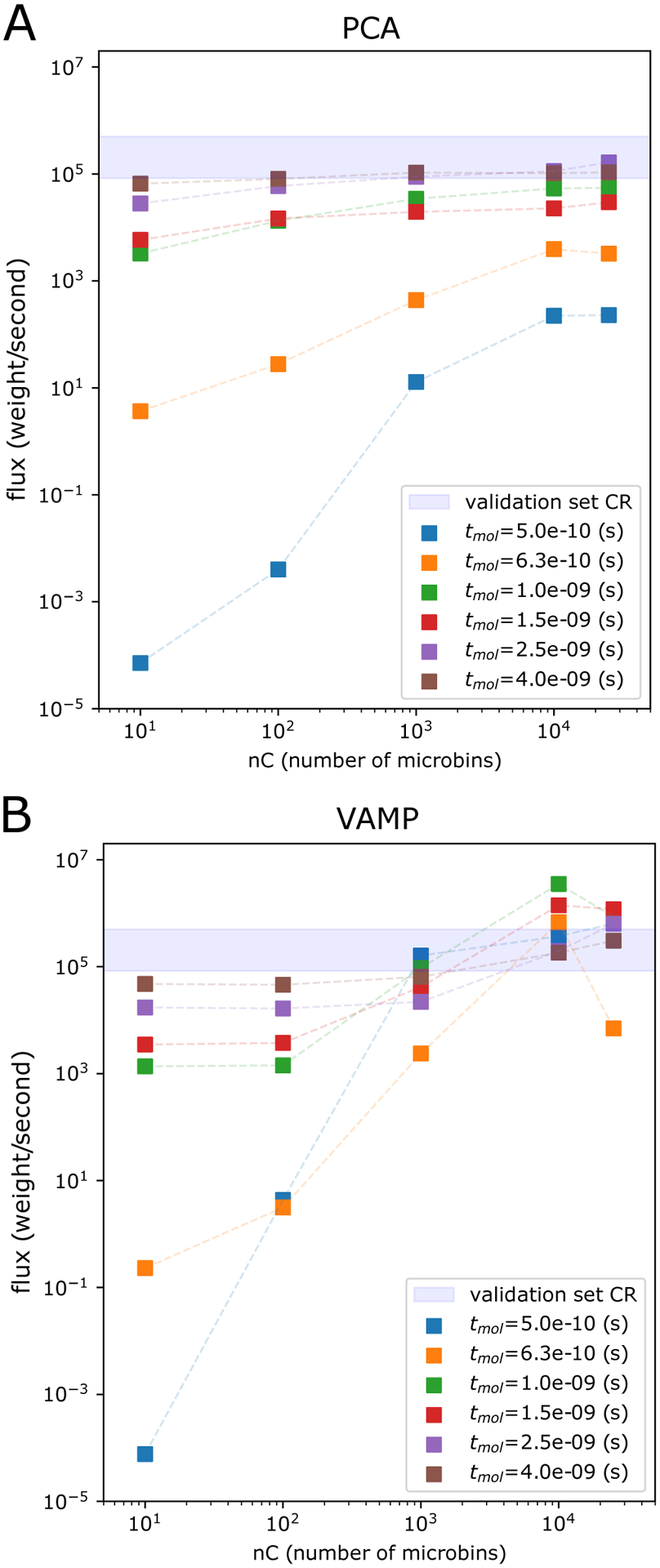
Steady state estimation for NTL9 protein-folding using haMSMs with PCA and VAMP microbins. Shown is the haMSM-estimated SS flux for NTL9 (low-friction) protein folding at tmol = 0.5, 0.63, 1.0, 1.5, 2.5, 4.0ns (colored squares) as a function of the number of haMSM microbins, and SS flux from WE validation set (shaded blue horizontal bar). A Principal Component Analysis (PCA) dimensionality reduction. B VAMP dimensionality reduction.
