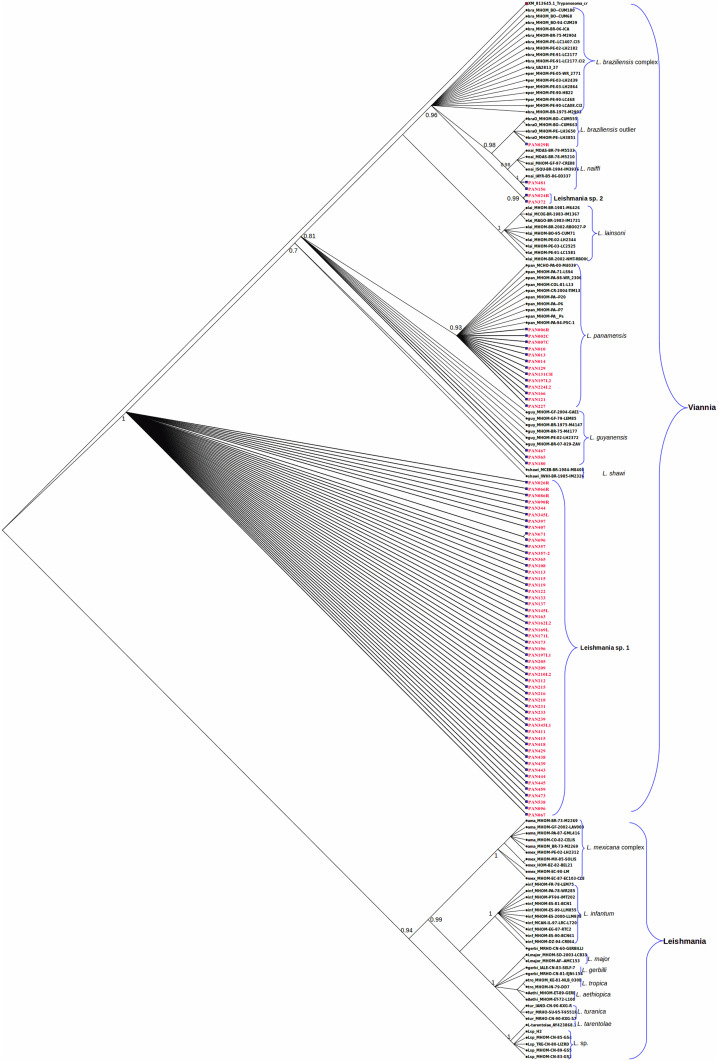
Figure 2.
Hsp70-based Bayesian phylogenetic tree of Leishmania species obtained constructed under the GTR (Nst = 6) model with a gamma rate of four. The Bayesian consensus tree was searched by the program Mr. Bayes after 8,000,000 cycles of the Markov Chain Monte Carlo algorithm with a subsampling frequency of 1,000. Clade credibility values are shown as values at each clade node. Reference sequence codes appear in black color. All sequence codes obtained in this study are highlighted in red color. Reference sequences are abbreviated as follows: bra: Leishmania braziliensis; braO: Leishmania braziliensis outlier; guy: Leishmania guyanensis; lai: Leishmania lainsoni; nai: Leishmania naiffi; pan: Leishmania panamensis; per: Leishmania peruviana. This figure appears in color at www.ajtmh.org.