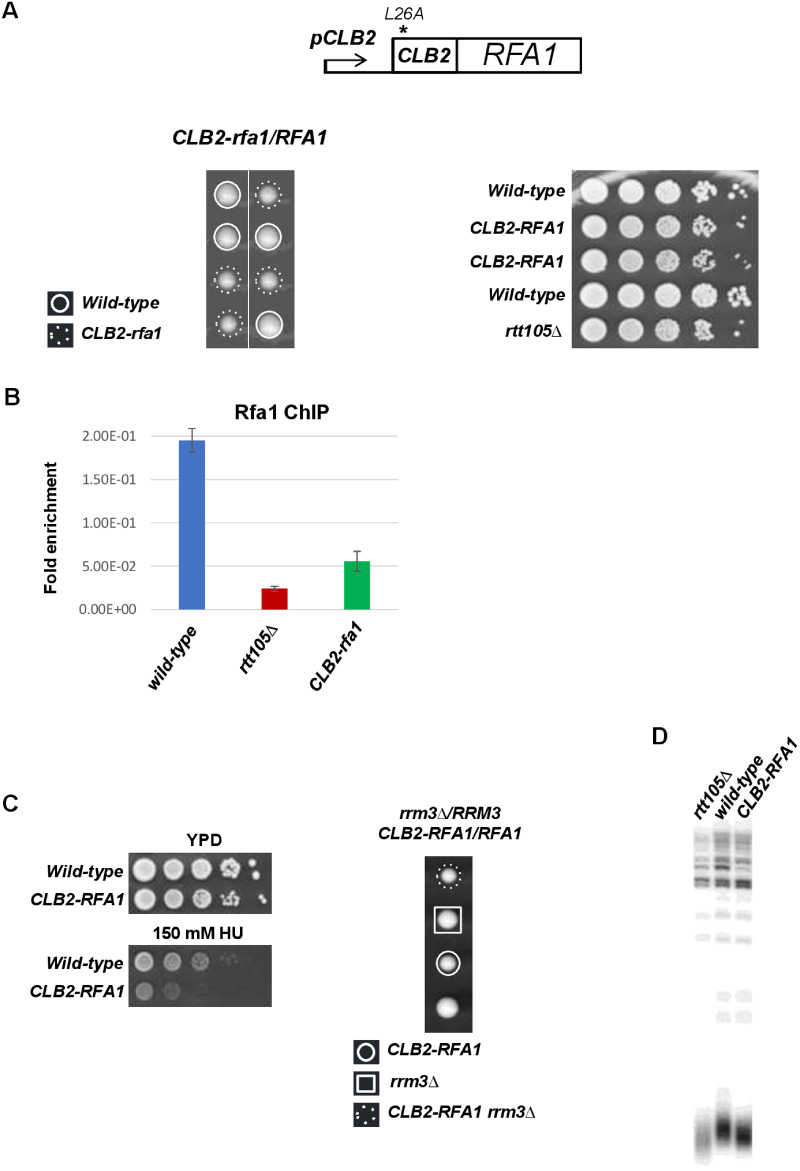
Figure 8.
Phenotypes of CLB2-rfa1 cells. (A) Top, schematic representation of the CLB2-rfa1 construct used in this study. Down, RTT105 is required for normal cell growth. Left, tetrad dissection of the diploid strain CLB2-rfa1/RFA1. Right, 10-fold serial dilutions of exponentially growing haploid cells were spotted. (B) Reduced binding of Rfa1 at fork in CLB2-rfa1 cells. Asynchronous cells were blocked in S-phase with 200 mM HU. ChIP experiments were performed in triplicate using an antibody against Rfa1 (Agrisera) and the resulting DNA was quantified with real time PCR using primers amplifying ARS607 (CGTGCGGCAGTATAAGTTCA and GCAGGATCGACCTGACTCTT). (C) CLB2-rfa1 mutant viability is affected by HU but not by RRM3 inactivation. Left, 10-fold serial dilutions of exponentially growing cells were spotted onto YPD plate or 150 mM HU plate. Right, tetrad dissection of the diploid strain CLB2-rfa1/RFA1 rrm3Δ/RRM3. Plates were incubated at 30°C for 3 days. (D) Effect of CLB2-rfa1 on telomere length measured by Southern blotting.