Figure 6.
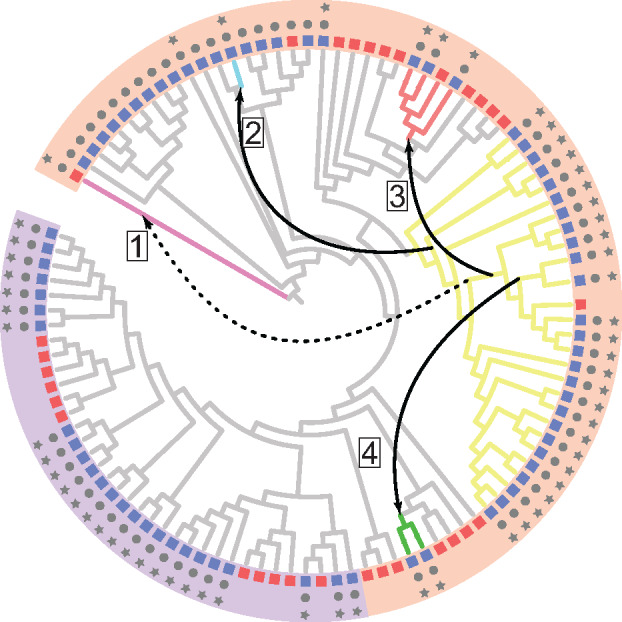
The CUG-Ser1 clade serves as a common donor of the GAL gene cluster to other yeasts. A cladogram of the ML phylogeny is presented with the leaf labels removed for simplicity. The colored boxes represent the species’ ability to utilize galactose (blue = positive/variable, red = negative), gray circles indicate the presence of a full set of GAL enzymatic genes, and gray stars indicate that those GAL genes are clustered. Five lineages on the cladogram are colored: pink—Sc. pombe (a member of the subphylum Taphrinomycotina with a transferred GAL cluster that does not confer utilization), light blue—Nadsonia, red—Brettanomyces, yellow—CUG-Ser1 clade, and green—Wickerhamomyces. Numbered boxes and arrows depict the four horizontal transfer events of the GAL cluster, one previously documented case into Sc. pombe (dashed arrow) (Slot and Rokas 2010) and the three described here (solid arrows). The colored arcs encompassing the cladogram represent the predicted regulatory mode of the GAL genes: orange—Rtg1/Rtg3 (non-Gal4) and purple—Gal4.
