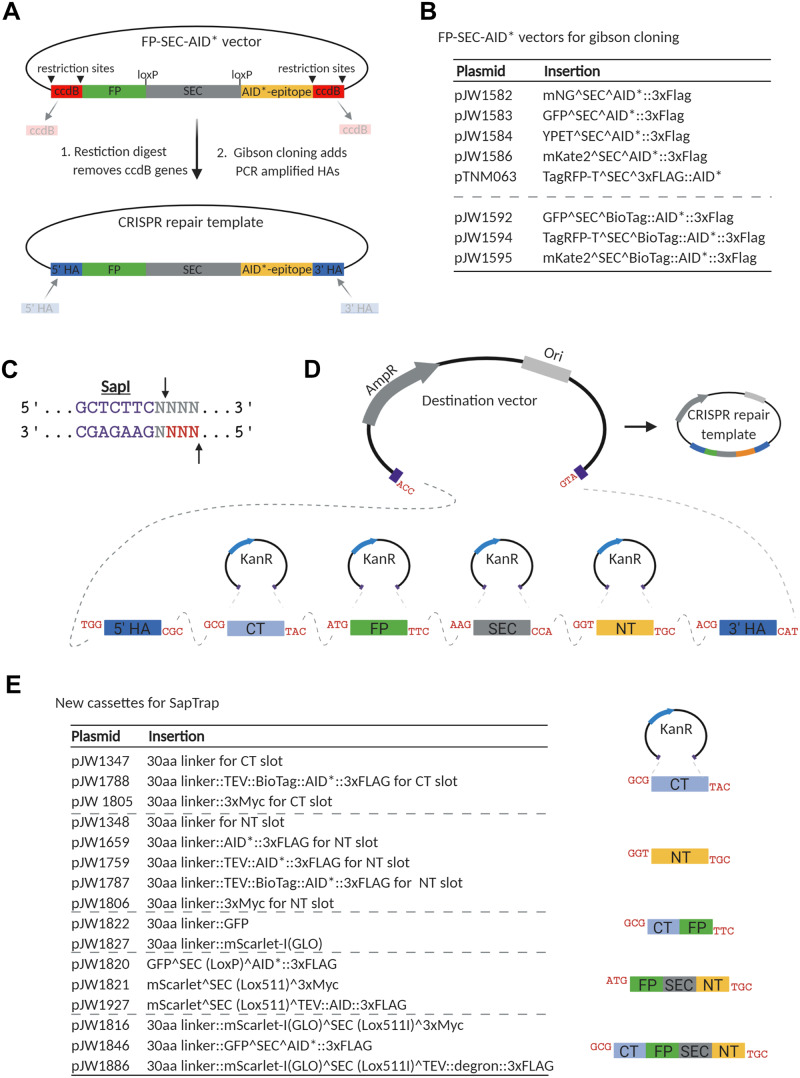
Figure 8.
A collection of vectors to generate FP::AID* knock-ins through Gibson cloning and a suite of new vectors for the SapTrap cloning system. (A) Schematic of the AID*-containing vectors produced by modifying the set of vectors originally described by Dickinson et al. (2015). An AID* epitope was inserted downstream of the loxP-flanked SEC. New repair templates for CRISPR/Cas9-mediated genome editing can be produced by restriction digestion of the vector and Gibson cloning of PCR-derived 5’ and 3’ homology arms (5’HA and 3’HA), as described (Dickinson et al. 2015). Counter-selection against the parent vector is provided by ccdB cassettes. Created with BioRender.com. (B) A suite of new FP::AID* SEC plasmids. The vectors described in Dickinson et al. (2015) have been modified to insert an AID* or 23 amino acid biotin acceptor peptide (BioTag)::AID* cassette between the SEC and 3xFLAG cassette. (C) SapI is a type II restriction enzyme that cuts one base pair and four base pairs outside of its binding site, allowing for the generation of programable 3 bp sticky ends. D) SapTrap cloning facilitates single-reaction cloning of multiple fragments, in the correct order, into a single repair template plasmid. Specific sticky ends are used for specific cassettes as described by Schwartz et al. (2016). Created with BioRender.com. E) Table of new vectors generated for the SapTrap CT and NT slots. Our initial assembly efficiencies were sub-optimal, and we found that reducing the number of fragments assembled improved our efficiencies. We have generated a set of multi-cassettes where partial assemblies (CT-FP, FP-SEC-NT, and CT-FP-SEC-NT) have been cloned, simplifying the SapTrap reactions and reducing the number of fragments required. 5’HA, 5’ homology arm; 3’HA, 3’ homology arm; FP, fluorescent protein; SEC, self-excising cassette; CT, C-terminal connector; NT, N-terminal connector.