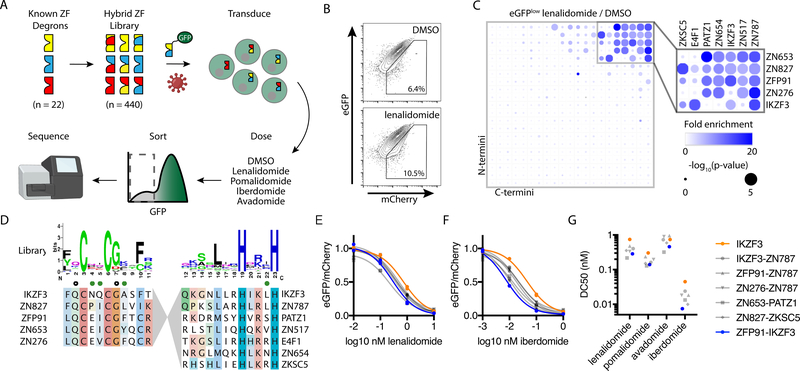
Fig. 3: A screen of 440 hybrid zinc fingers identifies “super-degrons” targeted by subnanomolar concentrations of thalidomide analogs.
(A) Schematic for the design and screening of a hybrid zinc finger (ZF) library encoded in a eGFP-tagged protein degradation reporter lentivector that also expressed mCherry as a control for lentivector transgene expression. Jurkat cells were transduced with the lentivirus library and then exposed to various thalidomide analogs or vehicle control (DMSO). FACS was used to isolate eGFPlow cells, and next-generation sequencing (NGS) was used to quantify the relative abundance of each sequence with and without drug treatment. (B) Flow plot for Jurkat cells transduced with the eGFP-tagged zinc finger library after overnight incubation with 1 μM lenalidomide or vehicle control. (C) Fold-enrichment of sequencing read counts (lenalidomide/DMSO, scale 0 – 20) and corresponding empirical rank-sum test P values (0 – 5). Average of nine replicates is presented (library of triplicate barcoded sequences assayed with triplicate independent biologic replicates). (D) Sequence features for N- and C-terminal domains present in top candidate super-degrons. Amino acid positions with prior crystallographic evidence of side-chain interactions with pomalidomide (open circle) or CRBN (green circle) are noted. Jurkat cells were engineered to express the indicated zinc finger constructs and treated with various thalidomide analogs. (E and F) Degradation of GFP-tagged zinc fingers was assessed by flow cytometry from the vehicle control-normalized eGFP/mCherry fluorescence ratio in Jurkat cells after treatment with lenalidomide (E) or iberdomide (F). Individual values and nonlinear regression are shown. Experiment was performed once in technical duplicate. (G) Nonlinear regression was used to calculate half maximal degradation concentration (DC50) values for zinc finger degradation by each drug.