Figure 2. Oncogenic RAS-dependent control of cellular metabolism.
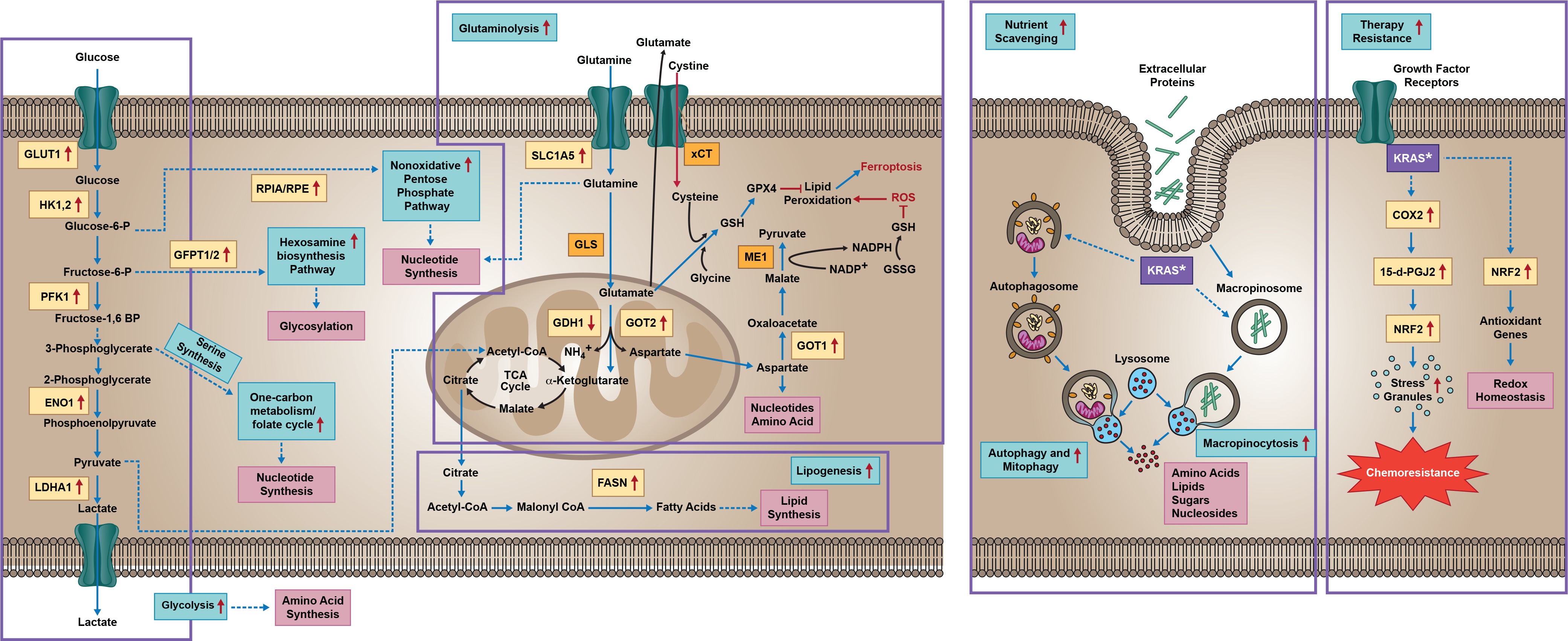
Mutant RAS deregulates key metabolic processes, including glutaminolysis, glycolysis, autophagy, and macropinocytosis. Oncogenic KRAS directs glucose metabolism into hexosamine biosynthetic pathways by upregulating several key glycolytic enzymes, and induces the nonoxidative pentose phosphate pathway to support increased nucleic acid biosynthesis. RAS-driven cancer cells alter glutaminolysis to support rewired metabolism. Altered glutaminolysis is a key feature of KRAS-dependent cancer cells. KRAS regulated glutamine metabolic rewiring influenced the TCA cycle, which is critical for nucleotide biosynthesis to support cell growth and survival. KRAS-driven tumors require glutaminolysis for redox balance. KRAS-mediated activated NRF2 is depicted as a key transcription factor that modulates redox homeostasis for the survival of cells under oxidative stress. Cells harboring mutant RAS are characterized by increased macropinocytosis, autophagy, and mitophagy, processes which help generate the energy and macromolecules needed for accelerated tumor proliferation. Mutant RAS also regulates stress granule formation, which helps mediate chemoresistance. Yellow box indicates RAS-dependent gene and/or protein expression, with arrows indicating increased or decreased expression. Purple box indicates oncogenic KRAS. GDH1: Glutamate DeHydrogenase 1; TCA: Tricarboxylic Acid; COX2: Cyclooxygenase 2; HK1,2: Hexokinase 1 and 2; GLUT1: Glucose Transporter-1; PFK1: Phosphofructokinase-1; ENO1: alpha-enolase-1; LDHA: Lactate Dehydrogenase; ME1: Malic Enzyme-1; ROS: Reactive Oxygen Species; GOT1,2: Glutamate Oxaloacetate Transaminase 1,2.
