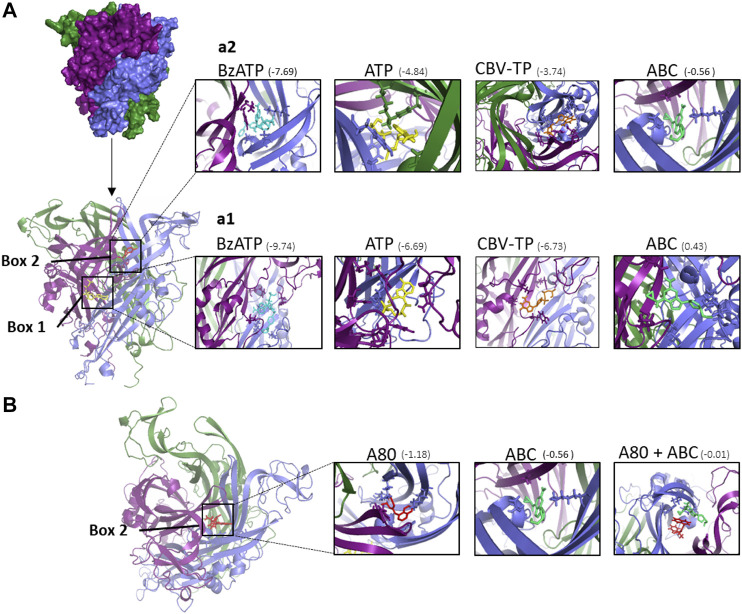
FIGURE 7.
Docking calculations of ABC and other ligands to P2X7 receptor. (A) Structure of human P2X7R built by homology modeling based on the X-ray crystal structure of panda P2X7R (5U2H). The P2X7R trimer is represented as a molecular surface. The three monomers are colored in deep purple, slate blue, and forest green. Each monomer is shaped like a dolphin (Hattori and Gouaux, 2012). Docked poses at ATP binding site (box 1) and A804598 (A80) binding site (box 2) of the different compounds studied. The P2X7R trimer is represented as a cartoon. ATP is depicted in yellow, A804598 in red, Bz-ATP in blue and CBV-TP in orange and ABC in lime green. (a1) Details of the ligands docked at box 1 showing some amino acid side chains at the interaction site. (a2) Details of the ligands docked at box 2 showing some amino acid side chain at the interaction site. (B) Superimposition of docked poses of P2X7R antagonist A804598 (A80, in red) and ABC (in lime green) at the binding site two of the P2X7R. The predicted binding energies are indicated under parenthesis (kcal mol−1).