FIGURE 1.
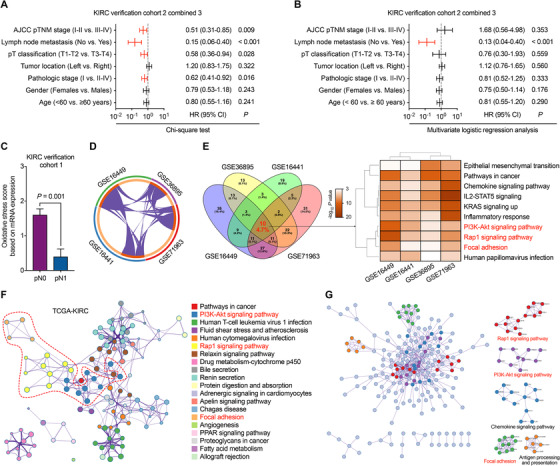
Correlation between oxidative stress score and KIRC patients’ clinicopathological features. A‐B, Univariate and multivariate analyses between clinicopathological characteristics and oxidative stress score in verification KIRC cohort 2 combined 3. C, oxidative stress score calculated in KIRC verification cohort 1 grouped by pN stage. Bar, SEM; Student's t‐test. D, Analysis of genes associated with oxidative stress score in 4 GEO‐KIRC databases. E, Pathway and process enrichment analyses of oxidative stress score‐related genes in 4 GEO‐KIRC databases. Terms with a P < 0.05, a minimum count of 3, and an enrichment factor > 1.5 are collected and grouped into clusters based on their membership similarities. F, Top 20 network of enriched terms associated with oxidative stress score in the TCGA‐KIRC database. Terms with a similarity > 0.3 are connected by edges. G, Protein‐protein interaction enrichment analysis of oxidative stress score‐related genes in TCGA‐KIRC database.
Abbreviations: KIRC, kidney renal clear cell carcinoma; AJCC, American Joint Committee on Cancer; HR, hazard ratio; CI, confidence interval; IL2‐STAT5, interleukin 2‐signal transducer and activator of transcription 5; KRAS, Kirsten rat sarcoma 2 viral oncogene homolog; PI3K, phosphoinositide 3‐Kinase; PPAR, peroxisome proliferator‐activated receptor; GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas.
