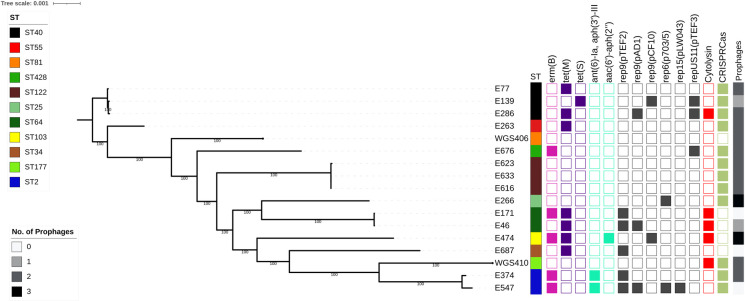
Fig 1. Core genome phylogeny of E. faecalis endophthalmitis isolates.
Single-copy core genome phylogeny of 15 UPMC Campbell Lab isolates plus two additional genomes from NCBI (WGS406 and WGS410), all isolated from endophthalmitis. The RAxML tree is built from single nucleotide polymorphisms (SNPs) in 2024 single copy core genes identified with Roary. Tips are annotated with isolate name, multi-locus sequence type (ST), drug resistance-associated genes, plasmid rep genes, cytolysin operon presence, CRISPR-cas loci presence, and prophage abundance in each genome. CRISR-cas-positive isolates had Class 1 and/or Class 2 Cas proteins associated with their CRISPR loci. Intact or questionable prophages were identified with PHASTER. erm(B) = erythromycin resistance, tet(M) and tet(S) = tetracycline resistance, ant(6)-Ia, aph(3′)-III and aac(6’)-aph(2″) = aminoglycoside resistance.