Fig. 1. Kog1 controls growth under glucose and amino acid limitation.
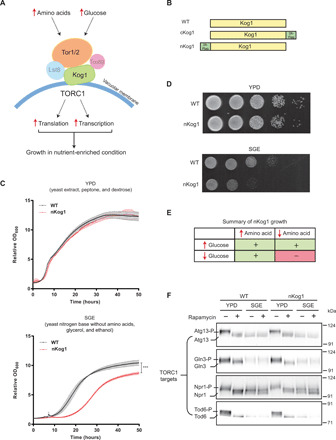
(A) A schematic showing components of the TORC1 pathway. Kog1 is a central scaffold that holds the different components of TORC1 together. (B) A schematic indicating the Kog1 strains used in this study. Kog1 was chromosomally tagged at C or N termini with a 3X-Flag epitope and named cKog1 and nKog1, respectively. WT is the unaltered, wild-type strain. (C and D) nKog1 cells show a growth defect in minimal medium with glycerol and ethanol as the carbon source (SGE). Growth curves and serial dilution spot assay for WT and nKog1 cells in YPD and SGE medium. Data are represented as means ± SD (n = 2). ***P < 0.0001 (unpaired Student’s t test). Also see fig. S1 (A to D) for growth in other conditions and fig. S1E for the growth of ∆kog1. (E) Summary of the comparative growth of nKog1 versus WT cells in different nutrient combinations (variables: glucose, glycerol/ethanol, free amino acids). “+” indicates normal growth and “−” indicates reduced growth compared to WT. (F) Canonical TOR kinase outputs are unaffected in nKog1. Western blots showing the electrophoretic mobility of Tor kinase targets—Atg13, Gln3, Npr1, and Tod6—in WT and nKog1 cells, in YPD, and after a 1-hour shift to SGE medium. Respective proteins were epitope tagged with hemagglutinin (HA) and detected using anti-HA antibodies. A representative blot (n = 2) is shown. Also see fig. S1 (F to I) for Kog1 protein, association of Kog1 with Tor kinase, and Sch9 electrophoretic mobility.
