Fig. 6. Kog1 controls Snf1 activation to regulate growth under nutrient limitation.
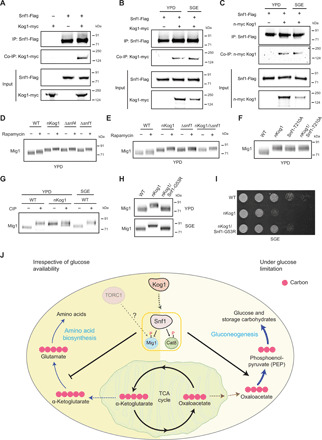
(A to C) Snf1 interacts with Kog1. Flag-tagged Snf1 was immunoprecipitated from cells in YPD or after a 1-hour shift to SGE medium and eluted, and coimmunoprecipitated myc-tagged Kog1 was detected. A representative image (n = 2) is shown. Also see fig. S6A. IP, immunoprecipitation. (D and E) A representative Western blot (n = 3) depicting the electrophoretic mobility of Mig1 in WT, nKog1, Δsnf4, Δsnf1, and nKog1/Δsnf4 cells before and after rapamycin treatment in YPD. Also see fig. S6 (B and C). (F) A representative Western blot (n = 2) depicting the electrophoretic mobility of Mig1 in WT, nKog1, Snf1-T210A, and nKog1/Snf1-T210A cells in YPD medium. (G) Cell lysates from WT and nKog1 cells in the indicated conditions were treated with phosphatase (CIP), and the electrophoretic mobility of Mig1 was assessed. A representative image (n = 3) is shown. Also see fig. S6D. (H) A representative Western blot (n = 2) depicting the electrophoretic mobility of Mig1 in WT, nKog1, and nKog1/Snf1-G53R cells in YPD and after a 30-min shift to SGE medium. (I) Comparative growth of WT, nKog1, and nKog1/Snf1-G53R cells in SGE medium. Also see fig. S6H. (J) A mechanistic model proposing Kog1-dependent regulation of SNF1. Kog1 controls SNF1 activation irrespective of nutrient availability, and reduces carbon flux toward amino acid biosynthesis. Upon glucose limitation, the Kog1-dependent activation of SNF1 (and phosphorylation of Mig1 and Cat8) regulates growth by appropriately inducing gluconeogenesis and increasing carbon flux toward the synthesis of gluconeogenic intermediates and sugars.
