Fig. 2. Crystallographic screening identified 234 fragments bound to Mac1.
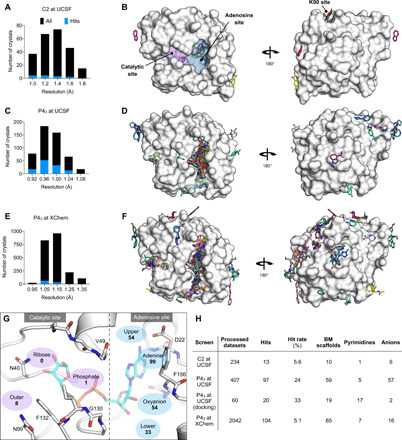
(A, C, and E) Histograms showing the resolution of the crystallographic fragment screening data. The resolution of datasets where fragments were identified are shown with blue bars. (B, D, and F) Surface representation of Mac1 with fragments shown as sticks. (G) The Mac1 active site can be divided on the basis of the interactions made with ADPr. The “catalytic” site recognizes the distal ribose and phosphate portion of the ADPr and harbors the catalytic residue Asn40 (10). The “adenosine” site recognizes adenine and the proximal ribose. The number of fragments binding in each site is indicated. (H) Summary of the fragments screened by x-ray crystallography, including the number of BM scaffolds and anionic fragments identified as hits in each screen. “Processed datasets” refers to the number of datasets that were analyzed for fragment binding with PanDDA. Of the datasets collected for 2954 fragments, 211 (7.1%) were not analyzed because of data pathologies.
