Abstract
Background
Small cell lung cancer (SCLC) is an aggressive and invasive malignancy that presents at advanced clinical stage with no more effective treatments. Development of a method for its early detection would be useful, also new therapeutic target need to be discovered; however, there is a lack of information about its oncogenic driver gene mutations.
Objectives
We aim to identify the SCLC-related genomic variants that associate with clinical staging and serum protein biomarkers observed in other types of lung cancer.
Methods
We screened formalin-fixed paraffin-embedded (FFPE) biopsy tissues of 32 Chinese SCLC patients using the 303 oncogenic driver gene panel generated by Tiling PCR amplification sequencing (tPAS) and analyzed the patients' corresponding serum protein levels of CYFRA21-1 CEA, NSE, and SCCA.
Results
In total, we found 147 SCLC-related mutant genes, among these, three important genes (TP53, RB1, KMT2D) as well as five novel genes LRRK2, BRCA1, PTCH1, ARID2, and APC that altogether occurred in 90% of patients. Furthermore, increased mutations to 6 genes (WT1, NOTCH1, EPHA3, KDM6A, SETD2, ACVR1B) significantly associated with higher serum NSE levels (P = 0.0016) and higher clinical stages II + III compared to stage I (P = 0.06).
Conclusions
Our panel is relatively reliable in detecting the oncogenic mutations of Chinese SCLC patients. Based on our findings, it may be possible to combine SCLC-related mutations and serum NSE for a simple detection of clinical staging.
1. Background
Lung cancer is a major cause of cancer-related deaths worldwide [1, 2]. Small cell lung cancer (SCLC), accounting for approximately 15% of lung cancers [3], is an aggressive, neuro-endocrine tumor characterized by a short doubling time, high growth rate, and early onset of metastases [4, 5]. While treatment includes surgery, chemotherapy and radiotherapy [6], the majority of patients present at advanced stages with loco-regional and systemic involvement and have poor prognosis and limited treatment options. Thus, a method for the early detection of SCLC is required and new treatments' targets need to be discovered.
There are no specific protein biomarkers identified for the SCLC. However, there are growing reports of a number of potentially useful serum proteins for the diagnosis of lung, prostate, and colorectal cancer [7–9]. Significantly, shorter survival rates have been associated with high levels of circulating tumor biomarkers, e.g., cytokeratin-19 fragment, neuron-specific enolase (NSE), and thymidine kinase, in patients with non-small-cell lung carcinoma (NSCLC) [10]. Tumor biomarkers can be further combined with certain mutations to improve the sensitivity for the detection of cancers, such as the carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9), osteopontin, and hepatocyte growth factor that are combined with KRAS mutation to detect pancreatic cancer [11].
Due to the low incidence and survival rates from SCLC [12], as well as difficulty in obtaining sufficient tissue samples for research [13], still not enough is known about the driver mutations that associate with cancer development and progression. So far, exome sequencing of SCLC has highlighted the important mutations in the TP53, RB1, and histone-modifying genes [14]. Inactivating mutations in TP53 and RB1 are found in 65-90% of SCLC patients [14–17]. Truncating frameshift, splice, and nonsense site variants in the KMT2D, which encodes a histone H3K4 methyltransferase, are found in 8% of SCLC tumors and 17% of SCLC cell lines [18].
To increase our understanding of the mutational signatures of SCLC and assist with development of new diagnostic tools, we analyzed the formalin fixed and paraffin embedded (FFPE) biopsy specimens of 32 Chinese SCLC patients with a new technology called Tiling PCR amplification sequencing (tPAS) based on the sequencing of multiplex PCR amplicons, which can simultaneously genotype single nucleotide variants (SNVs), insertions or deletions (indels), and copy number variations (CNVs) in a panel of 303 cancer genes, selected from the Catalogue of Somatic Mutations in Cancer [19], Cancer Genome Atlas [20, 21], and Oncomine database [22].
2. Materials and Methods
2.1. Study Participants
We recruited 32 patients diagnosed with SCLC into this study. The tumor specimens were obtained from patients by surgeons in the Oncology Department of The First Affiliated Hospital of Dalian Medical University. All participants gave written informed consent for the provision of clinical information, biospecimen collection, and analysis. This study was approved by the Dalian Medical University and Hospital Research Ethics Committee. The Institutional Review Board (IRB) for this study is YJ-JG-QX-2018-146.
2.2. Pathological Analysis of Tumor Specimens
Tumor specimens were acquired by surgery (>2% of total tissue mass and >150 cells). Diagnosis of SCLC was confirmed by pathologists using histologic evaluation of the stained FFPE tumor sections. TNM staging system of International Association for the Study of Lung Cancer (version 7) was used to determine the clinical staging.
2.3. Extraction of DNA from Formalin Fixed Paraffin Embedded (FFPE) Biopsy Specimens
Macrodissection was performed to increase the tumor tissue percentage to ~80% prior to DNA extraction. Five to ten FFPE sections (5 mm thickness) of each tumor specimen were used to prepare the genomic DNA for mutation profiling. DNA was extracted from FFPE tissue using the AmoyDxò FPPE DNA Kit and DNA purification spin columns (Amoy, Xiamen, Fujian). All purified DNA samples were determined to be of high quality by spectroscopy analysis and were suitable for mutation analysis by the tPAS assay.
2.4. Targeted Next-Generation Sequencing (NGS)
Targeted NGS was performed on the 32 FFPE samples. Libraries were prepared according to Paragon Genomics manufacturer's protocol. Briefly, 40 ng of human DNA was used for each multiplex PCR reaction. CleanPlex™ panels were supplied with 2x concentrated primer pools. Using thin-wall PCR strip tubes, components were added to run the PCR program for amplify target DNA regions. For panels consisting of multiple primer pools, 10 μl multiplex PCR reactions were combined for each sample. Magnetic bead suspensions were vortexed vigorously, and 1.3x sample volume of magnetic bead suspension was added to each sample. After beads were drawn onto one side of the wall, the supernatant was removed and discarded without touching the beads, 10 μl TE buffer was added to each tube, and then, the DNA was immediately released from the beads. Amplicons were purified, and concentration of the library was measured.
2.5. Tiling PCR Amplification Sequencing (tPAS)
Paired-end 2 × 150 base pairs (bp) reads were generated from the Amplicon libraries using NextSeq CN500. Adaptor and low-quality bases in raw reads were trimmed using flexbar version v2.5 and aligned to the human reference genome hg19 using BWA-MEM version 0.7.5a. GATK v3.6 was used for local indel realignment and base quality recalibration [23]. GATK MuTect2 was used in somatic mutation (single nucleotide variants and small insertions and deletions) calling [24]. Somatic variant loci were defined by loci that were covered by ≥100 total reads and supported by ≥4 variant reads in the tumor and in the matched normal sample covered by ≥50 total reads and supported by ≤2 variant reads. ANNOVAR and TransVar were used for annotation with public variant databases [25, 26]. Variants were filtered if the baseline population frequency ≥ 5%.
2.6. Statistical Analysis
Nonparametric statistical tests were used throughout analysis due to a small sample size, and where P values were determined, the significance level was set at 0.05. Data for qualitative variables were reported as median and range. The association of clinical stage with clinical values, i.e., age and serum levels of protein biomarker, was analyzed as a continuous variable using the Wilcoxon rank sum test, while sex was analyzed as a categorial variable using the Fisher's exact test. The association between NSE levels and mutation counts was assessed using Spearman's correlation test. Statistical analysis was performed using R version 3.6.0.
3. Results
3.1. Baseline Characteristics
Table 1 shows the clinical characteristics of 32 Chinese SCLC patients. They tended to be older, at 65 years of age, and male. Under TNM classification, T2 and N0 were the most common categories (Table S1). We further classified patients into two broad clinical staging groups, stage I (n = 16) versus stages II + III (n = 16), to examine whether there were statistical differences between cancer stage and clinical values. Among the four investigated serum protein biomarkers, we found that serum levels of NSE of patients at stages II + III were significantly higher compared to stage I (P = 7 × 10–4; Table 1 and Figure S1).
Table 1.
Clinical characteristics of SCLC patients.
| Parameters | Clinical values∗ | Stage I | Stages II + III | P value∗∗ |
|---|---|---|---|---|
| Age (years old) | 65 (49, 81) | 67 (51, 81) | 63 (49, 81) | 0.2506 |
| Sex (male/female) | 24/8 | 12/4 | 12/4 | 0.9999 |
| CYFRA21-1 (ng/ml) | 4.94 (1.24, 19.53) | 3.41 (1.24, 19.53) | 3.07 (2.04, 4.43) | 0.5033 |
| CEA (ng/ml) | 4.33 (0.83, 18.37) | 2.97 (0.83, 16.59) | 2.52 (1.37, 18.37) | 0.9786 |
| NSE (ng/ml) | 14.37 (1.83, 34.00) | 10.74 (1.83, 20.26) | 15.60 (11.38, 34.00) | 7.56 × 10–4 |
| SCCA (ng/ml) | 0.97 (0.20, 3.08) | 0.88 (0.20, 1.83) | 0.86 (0.21, 3.08) | 0.9742 |
∗Reported are median and range, except for sex is total individuals. ∗∗P values for age, CYFRA21-1, CEA, NSE, and SCCA were generated by Wilcoxon rank sum test; P value for sex was generated by Fisher's exact test.
3.2. Identification of SCLC-Related Oncogenic Genes
An overview of this descriptive case study is shown in Figure 1(a). From the 32 SCLC patients, we detected a total of 147 oncogenes. Each patient harbored an average of 12.6 (range: 3, 90) pathogenic mutations. Total mutations and genes across patients arranged by clinical stage are shown in Figure 1(b). Table S2 shows the list of 147 SCLC-related genes ranked by total mutation counts. After we ranked the genes according to mutation number, we found TP53, RB1, and KMT2D were the most frequently mutated genes (representing top 2% of all genes, observed in 79% of patients). Including five more genes that shared same number of mutations, LRRK2, BRCA1, PTCH1, ARID2, and APC, increased the representation to top 5% of all genes, observed in 90% of patients. Together, these eight genes were the cut-off for the commonly mutated genes in this Chinese population of SCLC patients (Figure 1(c)).
Figure 1.
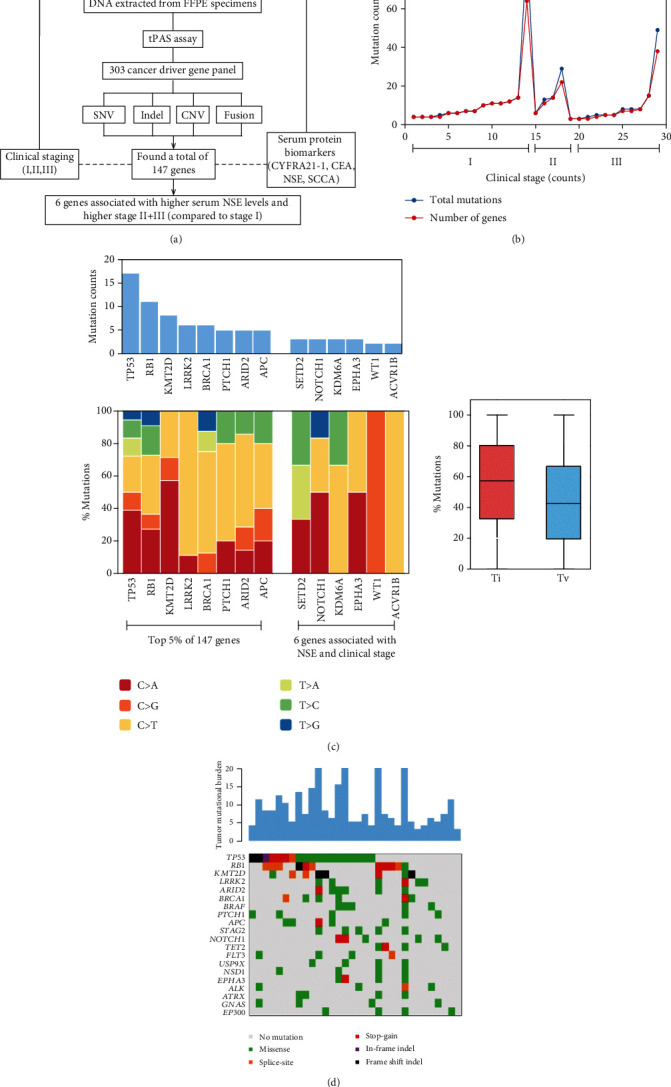
Study overview: (a) sequencing and analytical pipeline; (b) total mutations and number of genes per patient arranged according to clinical stages I, II, and III; (c) mutation counts of top 5% SCLC-related mutated genes observed in 90% of patients and 6 genes associated with NSE and clinical stage, where mutation to same gene is counted once (top) with their individual and combined summary of transition (Ti) and transversion (Tv) mutations (bottom); (d) mutational landscape of SCLC showing the number of coding somatic mutations per megabase of DNA (top) and a matrix of 20 frequently mutated genes colored by the type of variation (bottom).
3.3. Profiling of SNVs, Indels, and CNVs
The most common nucleotide mutations to the genes TP53, RB1, and KMT2D were C > A, C > T, and C > A transitions, respectively (Figure 1(c)). Total number of mutations per coding area of a tumor genome (tumor mutational burden (TMB)) ranged between 3 and 20 (Figure 1(d)). The mutational spectra of SCLC were dominated by missense, where indel and frameshift mutations occurred predominantly in TP53, RB1, and KMT2D. There were two mutation hotspots, c.832C > T (p.P278S) and c.1548G > A (p.W516X), observed in TP53 and RB1, respectively (Table S2). No CNV mutations were found in any of the genes.
3.4. SCLC Gene Variant Association with Serum Biomarkers and Clinical Stage
Because NSE associated with clinical stage, we also searched among the 147 genes for those that associated with NSE levels. We found 6 genes (WT1, NOTCH1, EPHA3, KDM6A, SETD2, ACVR1B) associated with serum NSE levels (P = 0.007–0.046; Figure 2). Generally, the more mutations to these 6 genes, the higher were the NSE levels (P = 0.0016) compared with one or no mutation to these 6 genes that were not associated with NSE levels (P = 0.1139), and furthermore, these 6 genes were associated with clinical stages II + III compared to I (marginal P = 0.06, Figure 3).
Figure 2.
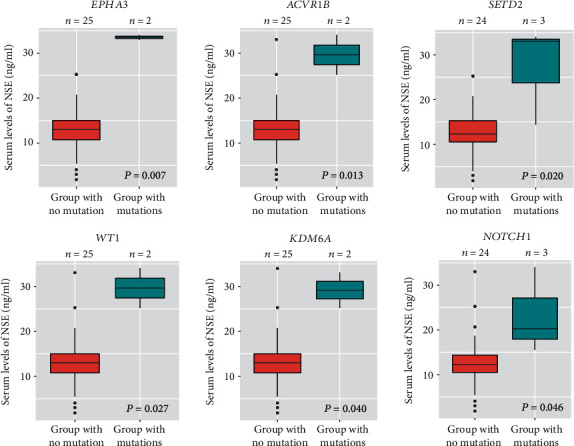
Significantly higher serum levels of NSE in SCLC patients with mutations to the following 6 genes (Wilcoxon rank sum test P = 0.007–0.046).
Figure 3.
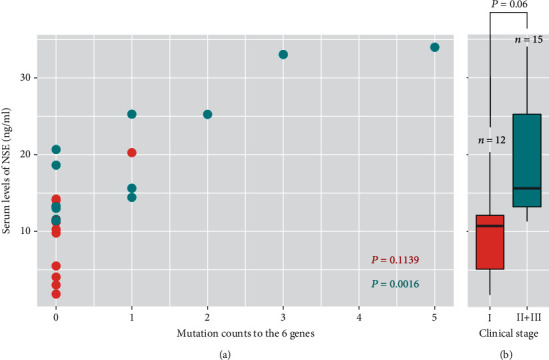
Increased mutations to 6 genes are associated with higher serum levels of NSE and clinical stage of SCLC. Scatterplot (a) shows patients with increased mutations to the following 6 genes (WT1, NOTCH1, EPHA3, KDM6A, SETD2, ACVR1B) have significantly higher NSE levels (Spearman's correlation test P = 0.0016) compared to those with ≤1 mutation to these 6 genes (P = 0.1139). Boxplot (b) shows patients with increased mutations to these 6 genes also have a significantly higher clinical stages II + III compared to stage I (Wilcoxon rank sum test P = 0.06).
4. Discussion
From 32 Chinese SCLC patients, we detected 147 out of 303 cancer driver oncogenes, including three previously implicated genes, TP53, RB1, and KMT2D, that appeared in 79% of our patients across all clinical stage. Together with LRRK2, BRCA1, PTCH1, ARID2, and APC, these eight genes represented top 5% of all genes that occurred in 90% of patients. In this study, we further found that serum protein marker NSE significantly associated with higher clinical stages II + III (P = 7 × 10–4). In particular, increased mutations to 6 genes (WT1, NOTCH1, EPHA3, KDM6A, SETD2, ACVR1B) significantly associated with higher NSE levels (P = 0.0016) and stages II + III (P = 0.06).
Loss of functions to TP53 and RB1 highly associate with the pathogenesis of SCLC [27] and are found in 82% and 62% of SCLC patients, respectively [28]. Previous study indicates that G:C to T:A transversions are the most frequently observed nucleotide substitutions to TP53 in lung cancer [29], which is consistent with our finding. In contrast, KMT2D mutations are less related to TP53 and RB1; it has instead been linked to a longer survival in patients with lung neuroendocrine tumors [30] despite that it has also been shown not to be involved in reducing survival of NSCLC patients [31]. In our study, the third most frequent gene mutations were in the KMT2D gene, whether the KMT2D gene plays a crucial role in survival of Chinese SCLC patient needs to be further investigated.
Among the five novel SCLC-related mutated genes found in this study, the LRRK2 is a mutation gene in the Chinese population with familial Parkinson's disease [32]. LRRK2 mutation carriers have an increased risk of cancer, especially for hormone-related cancer, and SCLC is associated with polypeptide hormone production [33]. The BRCA1 is involved in tumor suppression and homologous recombination repair in response to DNA breaks and may be a modulator of mitotic spindle assembly [34, 35]. Whole exome sequencing of the lung cancer has revealed a germline BRCA1 deficiency mutational signature [36]. We have identified mutations in LRRK2 and BRCA1 genes, and this represents the first report in the Chinese patients with SCLC. Finally, ARID2 is a tumor suppressor gene, where inactivating mutations along the ARID2 coding region is also detected in NSCLC [37]. ARID2 knockout results in dysfunction of DNA repair process, leading to susceptibility to carcinogens in human hepatocellular carcinoma cells [38]. However, it is unclear how ARID2 mutations are associated with SCLC progression. We identified a 46.9% variant allele fraction to c.3004A > G (p.T1002A) in ARID2 of a SCLC patient. To our knowledge, our data is the first to report such high frequency SNV of ARID2 gene in SCLC.
In this study, serum levels of NSE associated with higher clinical stage of SCLC. It has been reported that NSE (81.2%) has the highest sensitivity, followed by CEA (42.7%), CYFRA21-1 (32.3%), and SCCA (1%), and the average concentration of NSE was statistically higher in the patients with extensive disease (88.2%) compared to limited disease (73.3%) [39]. Recent studies, such as CancerSEEK, used 16 gene mutations combined with abnormal levels of 8 protein biomarkers for early diagnosis of lung cancer [40]. Similarly, we found 6 mutated genes that could associate with serum NSE to provide information about the severity of SCLC.
Some limitations of this study should be noted. First, we focused on SNV and indels as their function could be predicted by numerous methods and that heterogeneity has immediate clinical consequences for treatment selection [41]. Second, we could not evaluate recurrent noncoding, copy-number, or epigenetic mutations because the functional prediction methods for them were not yet available. Third, we did not assess micrometastases that are not visible clinically, so could not exclude those mutations in yet undiscovered driver genes of metastases. Finally, the top 5% mutated genes did not overlap with the 6 genes associated with NSE and stage, might be the small sample size, or that 6 genes represented a subpopulation of SCLC patients.
5. Conclusion
About 50% of oncogenic driver mutations are detected by our panel, suggesting it is moderately enriched for SCLC detection. Apart from capturing the most important SCLC-related mutant genes reported in other studies, we further found 5 genes and increased mutations to 6 genes associated with higher NSE and clinical stage. More investigations are required to evaluate the possibility of combining these mutant genes with NSE into a fast and simple companion diagnostic kit for clinicians to detect SCLC stage.
Acknowledgments
The authors wish to thank Liu Shihong, Wang Lientu, and Chin Lihan for their contributions to this manuscript. This work was supported by grants from the National Natural Science Foundation of China (81774078, 81802886) and by grants from the Natural Science Foundation of Liaoning province, China (20180550693).
Data Availability
The data are available upon the authors' reasonable request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Figure S1: serum levels of NSE significantly associated with a higher clinical stages II + III of SCLC (Wilcoxon rank sum test P = 7 × 10–4). Table S1: clinical category IASLC T1–4 (any N) M0 or N0–N3 (any T) M0 SCLC. Table S2: list of 147 SCLC-related mutant genes ranked by total mutation counts.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2018. CA: a Cancer Journal for Clinicians. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.National Lung Screening Trial Research Team, Aberle D. R., Adams A. M., et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England Journal of Medicine. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp A., Bhosle J., Abdelraouf F., Popat S., O'Brien M., Yap T. A. Development of molecularly targeted agents and immunotherapies in small cell lung cancer. European Journal of Cancer. 2016;60:26–39. doi: 10.1016/j.ejca.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Waqar S. N., Morgensztern D. Treatment advances in small cell lung cancer (SCLC) Pharmacology & Therapeutics. 2017;180:16–23. doi: 10.1016/j.pharmthera.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Pietanza M. C., Byers L. A., Minna J. D., Rudin C. M. Small cell lung cancer: will recent progress lead to improved outcomes? Clinical Cancer Research. 2015;21(10):2244–2255. doi: 10.1158/1078-0432.CCR-14-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shepherd F. A., Ginsberg R. J., Feld R., Evans W. K., Johansen E. Surgical treatment for limited small-cell lung cancer: The University of Toronto Lung Oncology Group experience. The Journal of Thoracic and Cardiovascular Surgery. 1991;101(3):385–393. doi: 10.1016/S0022-5223(19)36720-0. [DOI] [PubMed] [Google Scholar]
- 7.Patz E. F., Jr., Campa M. J., Gottlin E. B., Kusmartseva I., Guan X. R., Herndon J. E., II Panel of serum biomarkers for the diagnosis of lung cancer. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology. 2007;25(35):5578–5583. doi: 10.1200/JCO.2007.13.5392. [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Shi T., Qian W. J., et al. The clinical impact of recent advances in LC-MS for cancer biomarker discovery and verification. Expert Review of Proteomics. 2016;13(1):99–114. doi: 10.1586/14789450.2016.1122529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liotta L. A., Petricoin E. F., 3rd The promise of proteomics. Clinical Advances in Hematology & Oncology. 2003;1(8):460–462. [PubMed] [Google Scholar]
- 10.Fiala O., Pesek M., Finek J., et al. Prognostic significance of serum tumor markers in patients with advanced-stage NSCLC treated with pemetrexed-based chemotherapy. Anticancer Research. 2016;36(1):461–466. [PubMed] [Google Scholar]
- 11.Cohen J. D., Javed A. A., Thoburn C., et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(38):10202–10207. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., He J. The development of targeted therapy in small cell lung cancer. Journal of Thoracic Disease. 2013;5(4):538–548. doi: 10.3978/j.issn.2072-1439.2013.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucchi M., Mussi A., Fontanini G., Faviana P., Ribechini A., Angeletti C. A. Small cell lung carcinoma (SCLC): the angiogenic phenomenon. European Journal of Cardio-Thoracic Surgery. 2002;21(6):1105–1110. doi: 10.1016/S1010-7940(02)00112-4. [DOI] [PubMed] [Google Scholar]
- 14.Peifer M., Fernández-Cuesta L., Sos M. L., et al. Integrative genome analyses identify key somatic driver mutations of small- cell lung cancer. Nature Genetics. 2012;44(10):1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi T., Nau M., Chiba I., et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246(4929):491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 16.Wistuba I. I., Gazdar A. F., Minna J. D. Molecular genetics of small cell lung carcinoma. Seminars in Oncology. 2001;28(2 Supplement 4):3–13. doi: 10.1016/S0093-7754(01)90072-7. [DOI] [PubMed] [Google Scholar]
- 17.Mori N., Yokota J., Akiyama T., et al. Variable mutations of the RB gene in small-cell lung carcinoma. Oncogene. 1990;5(11):1713–1717. [PubMed] [Google Scholar]
- 18.Augert A., Zhang Q., Bates B., et al. Small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance) Journal of Thoracic Oncology. 2017;12(4):704–713. doi: 10.1016/j.jtho.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes S. A., Beare D., Gunasekaran P., et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Research. 2015;43(D1):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Cancer Genome Atlas Research Network, Weinstein J. N., Collisson E. A., et al. The Cancer Genome Atlas pan-cancer analysis project. Nature Genetics. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K., Wang H. Cancer Genome Atlas pan-cancer analysis project. Zhongguo Fei Ai Za Zhi. 2015;18(4):219–223. doi: 10.3779/j.issn.1009-3419.2015.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye L., Li H., Zhang F., Lv T., Liu H., Song Y. Expression of KIF23 and its prognostic role in non-small cell lung cancer: analysis based on the data-mining of Oncomine. Zhongguo Fei Ai Za Zhi. 2017;20(12):822–826. doi: 10.3779/j.issn.1009-3419.2017.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A., Hanna M., Banks E., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K., Lawrence M. S., Carter S. L., et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38(16, article e164) doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W., Chen T., Chong Z., et al. TransVar: a multilevel variant annotator for precision genomics. Nature Methods. 2015;12(11):1002–1003. doi: 10.1038/nmeth.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George J., Lim J. S., Jang S. J., et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang L., Huang J., Higgs B. W., et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLoS Genetics. 2016;12(4, article e1005895) doi: 10.1371/journal.pgen.1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 30.Simbolo M., Mafficini A., Sikora K. O., et al. Lung neuroendocrine tumours: deep sequencing of the four World Health Organization histotypes reveals chromatin-remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. The Journal of Pathology. 2017;241(4):488–500. doi: 10.1002/path.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardeshir-Larijani F., Bhateja P., Lipka M. B., Sharma N., Fu P., Dowlati A. KMT2D mutation is associated with poor prognosis in non-small-cell lung cancer. Clinical Lung Cancer. 2018;19(4):e489–e501. doi: 10.1016/j.cllc.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Guo J. F., Nie L. L., et al. A novel LRRK2 mutation in a mainland Chinese patient with familial Parkinson’s disease. Neuroscience Letters. 2010;468(3):198–201. doi: 10.1016/j.neulet.2009.10.080. [DOI] [PubMed] [Google Scholar]
- 33.Havemann K., Luster W., Gropp C., Holle R. Peptide hormone production associated with small cell lung cancer. Recent Results in Cancer Research. 1985;97:65–76. doi: 10.1007/978-3-642-82372-5_7. [DOI] [PubMed] [Google Scholar]
- 34.Lotti L. V., Ottini L., D'Amico C., et al. Subcellular localization of the BRCA1 gene product in mitotic cells. Genes, Chromosomes & Cancer. 2002;35(3):193–203. doi: 10.1002/gcc.10105. [DOI] [PubMed] [Google Scholar]
- 35.Mullan P. B., Quinn J. E., Gilmore P. M., et al. BRCA1 and GADD45 mediated G2/M cell cycle arrest in response to antimicrotubule agents. Oncogene. 2001;20(43):6123–6131. doi: 10.1038/sj.onc.1204712. [DOI] [PubMed] [Google Scholar]
- 36.Cedrés S., Felip E., Cruz C., et al. Activity of HSP90 inhibiton in a metastatic lung cancer patient with a germline BRCA1 mutation. Journal of the National Cancer Institute. 2018;110(8):914–917. doi: 10.1093/jnci/djy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manceau G., Letouzé E., Guichard C., et al. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma. Journal International Du Cancer. 2013;132(9):2217–2221. doi: 10.1002/ijc.27900. [DOI] [PubMed] [Google Scholar]
- 38.Oba A., Shimada S., Akiyama Y., et al. ARID2 modulates DNA damage response in human hepatocellular carcinoma cells. Journal of Hepatology. 2017;66(5):942–951. doi: 10.1016/j.jhep.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 39.Molina R., Auge J. M., Escudero J. M., et al. Mucins CA 125, CA 19.9, CA 15.3 and TAG-72.3 as tumor markers in patients with lung cancer: comparison with CYFRA 21-1, CEA, SCC and NSE. Tumour Biology. 2008;29(6):371–380. doi: 10.1159/000181180. [DOI] [PubMed] [Google Scholar]
- 40.Cohen J. D., Li L., Wang Y., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogelstein B., Papadopoulos N., Velculescu V. E., Zhou S., Diaz L. A., Kinzler K. W. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: serum levels of NSE significantly associated with a higher clinical stages II + III of SCLC (Wilcoxon rank sum test P = 7 × 10–4). Table S1: clinical category IASLC T1–4 (any N) M0 or N0–N3 (any T) M0 SCLC. Table S2: list of 147 SCLC-related mutant genes ranked by total mutation counts.
Data Availability Statement
The data are available upon the authors' reasonable request.


